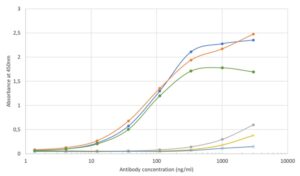
ELISA using anti-SARS-CoV-2 spike antibodies with the Omicron spike variant (B.1.1.529). ELISA plate was coated with the Omicron SARS-CoV-2 spike protein (REC32008) at 2.5 µg/ml. The antibodies and an isotype control were conjugated to HRP and titrated on a three-fold serial dilution starting at 3000 ng/ml. Controls and antibodies not recognizing the Omicron variant are shown at the bottom of the graph. Antibodies binding to the Omicron variant spike protein are shown at the top of the graph; MAB12422 is shown in blue.
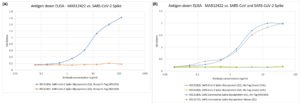
ELISA: Plate coated with the target proteins at 5 µg/ml. Primary antibodies were titrated on a 3-fold serial dilution starting at 125 ng/ml (A) or 41.6 ng/ml (B). Secondary antibody anti-human IgG conjugated to HRP used in the assay, at 1:4000 concentration. (A) Antibody recognised SARS-CoV-2 spike protein subunit 1 (aa 1-674), but not SARS-CoV-2 spike protein, subunit 2 (aa 685-1211). (B) Antibody recognised spike protein from SARS-CoV (subunit 1, aa 1-666) and SARS-CoV-2 (subunit 1, aa 1-674), produced in mammalian and insect cells, respectively. Antibody did not recognise SARS-CoV-2 spike protein, subunit 2 (aa 685-1211) or a spike mosaic protein, containing subunit 1 amino acids 12-53, 90-115, 171-203.
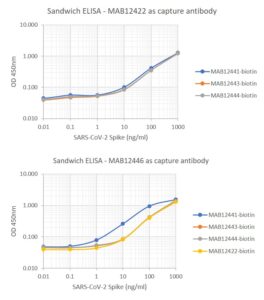
Sandwich ELISA: SARS-CoV-2 full-length Spike (REC31868) was the capture analyte. Plates were coated with 5µg/ml of capture antibodies. Spike protein was added at varying concentrations from 1µg/ml to 0.001ng/ml. Plates were incubated with biotin-labelled detection antibodies at 1µg/ml.
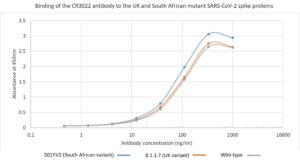
ELISA: ELISA using CR3022 antibody with UK (B.1.1.7) and South African (B.1.351 (501Y.V2)) mutant spike proteins (for mutant proteins see here). The plate was coated with mutant spike protein variants at 2.5 µg/ml. CR3022 was conjugated to HRP and titrated on a 3-fold serial dilution starting at 1,000 ng/ml. Strong binding was observed to all mutant spike proteins.
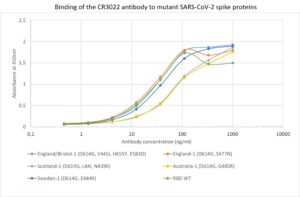
ELISA: ELISA using CR3022 and mutant spike proteins (for mutant proteins see here). The plate was coated with the mutant spike protein variants at 2.5 µg/ml. CR3022 was conjugated to HRP and titrated on a 3-fold serial dilution starting at 1,000 ng/ml. Strong binding to all mutant spike proteins was observed (RBD WT – wild-type receptor binding domain).
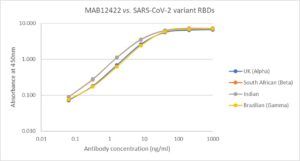
Direct ELISA: Plate was coated with 100µl of variant RBD proteins (UK, REC31946; South African, REC31945; Indian, REC31971; Brazilian, REC31961), at 1µg/ml and then incubated with 100µl MAB12422 antibody, diluted from 1000ng/ml to 0.064ng/ml. Diluted secondary IgG HRP antibody (100µl at 1:10,000) was then added. 100µl of TMB substrate (M0701A) was added in all wells and the reaction stopped after 10 min. with 1M HCl and the plate read at 405/450nm.
HUMAN IGG1 ANTI-SARS-COV-2 SPIKE (S1) ANTIBODY (CR3022)
Anti SARS-CoV-2 Spike (S1) antibody (CR3022) is a recombinant monoclonal antibody that recognizes the SARS-CoV and SARS-CoV-2 Spike glycoprotein, the causative agent of COVID-19. Antibody binds to both SARS-CoV and SARS-CoV-2 with high affinity at amino acids 318-510 (RBD, Receptor Binding Domain) in the S1 subunit of the Spike protein. Antibody recognizes RBD from Wuhan-Hu-1, UK (Alpha), South African (Beta) Brazilian (Gamma) and Indian variants. Suitable for use as a capture or detection antibody in ELISA assays.
PRODUCT DETAILS – HUMAN IGG1 ANTI-SARS-COV-2 SPIKE (S1) ANTIBODY (CR3022)
- This anti SARS-CoV-2 Spike IgG1 antibody binds the amino acids 318-510 in the S1 domain of the SARS-CoV Spike protein as well as SARS-CoV-2 (COVID-19) Spike protein.
- Isotype – human IgG1, Kappa.
- The original monoclonal antibody was generated by collecting peripheral blood lymphocytes of a patient exposed to the SARS-CoV.
- Protein A affinity purified from culture supernatant.
- Suitable for use in ELISA, NTRL, SPR, Crystallography.
- Suitable for use as a capture or detection antibody in ELISA assays.
BACKGROUND
Human coronaviruses are the major cause of upper respiratory tract illness. They are positive-stranded RNA viruses, and contain the largest viral RNA genomes known to date (27-31 kb). SARS (severe acute respiratory syndrome) and COVID-19 are both caused by human coronaviruses, SARS-CoV and SARS-CoV-2, respectively. The genome of SARS-CoV-2 shares 82% nucleotide identity with human SARS-CoV and 89% with bat SARS-like-CoVZXC21 (Lu et al., 2020; Zhao et al., 2020). However, initially it displayed lower pathogenicity and higher human to human transmissibility (Li et al., 2020). The coronavirus genome encodes four structural proteins: the spike (S) protein, nucleocapsid (N) protein, membrane (M) protein, and the envelope (E) protein. Cell entry is the first step in cross-species transmission and SARS-CoV-2 is likely to infect lung type II alveolar cells, which may explain the severe alveolar damage observed after infection (Zhao et al., 2020). Both SARS-CoV and SARS-CoV-2 uses the spike (S) protein to gain entry into host cells and it has been shown that the spike binds the entry receptor angiotensin-converting enzyme 2 (ACE2) on infected cells. It is predicted that SARS-CoV-2 recognizes human ACE2 more efficiently than SARS -CoV (Wan et al., 2020). Therefore, the S protein is considered a key target for vaccine development (Li et al, 2020).
This recombinant antibody binds to both SARS-CoV and SARS-CoV-2 (COVID-19) with high affinity; 86% of the epitope residues are conserved between SARS-CoV and SARS-CoV-2 (Tian et al., 2020; Yuan et al., 2020). The binding site is amino acids 318-510 (RBD, Receptor Binding Domain) in the S1 subunit of the Spike protein (Yuan et al., 2020). The antibody also binds to P462L-substituted S318–510 fragments of the SARS spike protein (Joyce et al. 2020). The crystal structure of this antibody has been resolved in complex with the RBD of the SARS-CoV-2 spike protein. The spike binding epitope is only accessible in the “open” conformation of the spike protein. The epitope can only be accessed by the antibody when at least two RBD on the trimeric spike protein are in the “up” conformation and slightly rotated (Joyce et al. 2020; Yuan et al., 2020). Antibody was originally isolated from a convalescent SARS patient, and is a neutralizing antibody that targets the receptor-binding domain (RBD) of SARS-CoV (ter Meulen, 2006). It did not neutralize SARS-CoV-2 in an in vitro assay, although it may show neutralisation in combination with other antibodies (Yuan et al., 2020). It has been shown to work synergistically in combination with antibodies that target the ACE2 binding site on the SARS-CoV RBD and reduce viral escape capacity. This antibody does not compete with ACE2 for binding to RBD, unlike most other known SARS RBD-targeted antibodies (ter Meulen, 2006). The epitope of this antibody does not overlap with the SARS-CoV-2 ACE2 binding site (Tian et al., 2020).
REFERENCES
- Joyce GM, et al. (2020). A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein. BioRxiv.
- Li Q, Guan X, Wu P, et al. (2020). Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382(13):1199–1207.
- Li H, Zhou Y, Zhang M, Wang H, Zhao Q, Liu J. (2020). Updated approaches against SARS-CoV-2. Antimicrob Agents Chemother. 2020;AAC.00483-20.
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574.
- ter Meulen J, van den Brink EN, Poon LL, et al. (2006). Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3(7):e237.
- Tian X, Li C, Huang A, et al. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382–385.
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol. 2020;94(7):e00127-20.
- Yuan M, Wu NC, Zhu X, et al. (2020). A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;eabb7269.
- Zhao et al. (2020). Single -cell RNA 2expression profiling of ACE2, the putative receptor of Wuhan 2019 -nCov. BioRxiv.
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273.