Since the emergence of SARS-CoV-2, scientists have speculated about the risks of mutation and what this could mean for therapeutics and vaccines. In this blog, we explore the nature viral mutation, what is known about the D614G mutant of SARS-CoV-2, and introduce our range of mutant Spike antigens.
Viral Variation
Mutation is the fundamental driver of life’s diversity. Simply put, mutations occur when the nucleotide sequences of genomes change. For viruses and other pathogens, mutations are typically of interest, as they can result in clinically-relevant traits, such as altered pathogenicity, transmissibility, host tropisms, drug resistance, and the ability to evade immune recognition. The extent to which viruses mutate and the way in which phenotypes manifest are therefore critical to understanding the origins, evolution and clinical outcomes of viral disease.
The propensity for a virus’s genome to mutate is influenced by a range of factors, such as its physio-chemical environment, genome structure, and accuracy of replication and repair mechanisms [1]. Some viruses are skilled molecular manipulators, actively seeking out opportunities generate mutations. Certain influenza viruses, for example, excel at genetic recombination and readily swap surface protein sequences with one another. The resulting collage of influenza subtypes can infect immunologically-naïve hosts with ease, requiring ongoing surveillance and annual reformulation of flu vaccines. HIV-1, on the other hand, makes use of mutation-inducing host enzymes, known as APOBECs, to accelerate resistance to drugs and evade immune recognition [3], and is one of the major reasons why HIV-1 countermeasures have been so difficult to develop.
However, while the very word “mutation” implies a substantial change, the biological reality is often much more benign [4]. In fact, the majority of viral mutations are of little-to-no consequence. Point mutations ― the most common type of mutation ― tend to result in either no change to the protein product, or substitution with functionally similar amino acids that do not significantly affect the phenotype. Moreover, more likely than conferring advantageous traits, mutations often impede essential functions, such as attachment, penetration, uncoating, replication, assembly, or release.
| |
|
|
| |
||
| |
||
| |
|
|
| |
||
| |
||
| |
||
| |
|
|
| |
Survival of the Flattest
The two most significant factors affecting the rate of viral mutation are the structure of a virus’s genome and the polymerase enzymes it uses to replicate this. DNA viruses (DNAVs), for example, typically have double‐stranded genomes, which they replicate using DNA-dependent DNA polymerases (DdDps) in host nuclei. RNA viruses (RNAVs), on the other hand, typically have single-stranded genomes which do not enter the nucleus, requiring them to encode their own RNA-dependent RNA polymerases (RdRps) for replication in the cytoplasm [5].
While such differences at first appear trivial, they can have drastic consequences. Firstly, single-stranded polynucleotides are less shielded from chemical and enzymatic attack from their surrounding environment. Secondly, because acute-infection RNAVs rely on rapid replication to outrun the host immune response, their RdRps must also work very quickly, and sacrifice the cumbersome proof-reading activity associated with polymerase complexes. This makes RdRps orders of magnitude less accurate than those of their hosts, or of DNAVs (by up to a million times [6]).
As a result of these factors, RNAVs typically incur significantly higher mutation rates than DNAVs. RNAV mutation rates are so high in fact, that many viral populations exist as ‘swarms’ of heterogenous mutants, known as quasispecies. Unlike the traditional paradigm of homogeneous viral populations, quasispecies have access to millions of sequence combinations that are able to complement one another [7]. This markedly changes the behaviour of RNAV populations and turns some Darwinian notions of evolutionary fitness on their head. Initially disadvantageous mutations, for example, can easily be sustained by fitter members of the population to facilitate more rapid adaptation to new environments. There’s even evidence to suggest that minority variants can constitute a form of ‘memory’ of past events that the population has experienced [8].
The benefit that low-fitness mutants confer is known as “survival of the flattest”, owing to the spread of fitness profiles across sequence space. The advantage of such mutations is explained by the fact that multiple nucleotide changes are typically required in sequence to arrive at a more advantageous genotype. Therefore, if certain mutations already exist, evolving to the new genotype is fewer steps away [9].
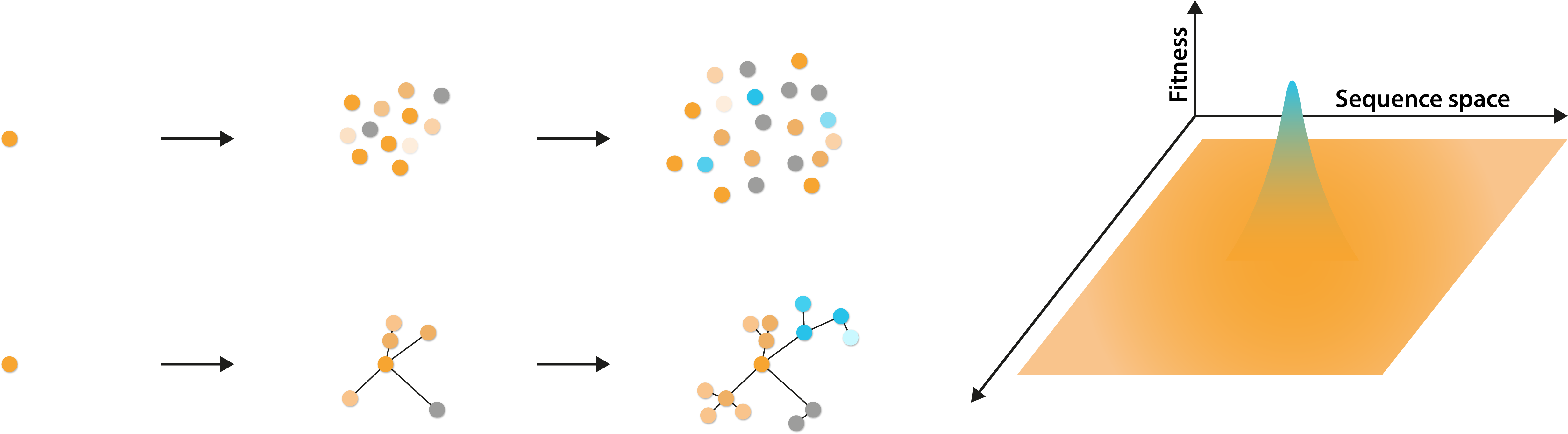
Left: Diagram illustrating the generation of a genetically diverse quasispecies from a single initial virus and their genetic relationship. Right: Diagram illustrating the potential fitness profiles of viruses, spread over a broad sequence space.
However, while a high rate of mutation appears to be a considerable advantage, it is not without its flaws. As many mutations are lethal, or at least highly disadvantageous, fluctuations in mutation rate can push viral populations perilously close to extinction (known as error catastrophe). Surprisingly, many RNAVs throw caution to the wind and operate close to their error catastrophe thresholds, regardless [10]. They manage this choice of lifestyle with a series of trade-offs, the most substantial of which, is the maintenance of small (<15kb) genomes that reduce the risk of accumulating too many deleterious mutations [11].
However, as RNAV genomes are constrained in size, they are inevitably limited in their potential complexity. However, as replication fidelity, genome size and genome complexity are highly interrelated, an increase in any one of these variables is inevitably prevented by the others. This conundrum is known as the Eigen trap, after German chemist, Manfred Eigen [11]. Since its inception, the Eigen trap has posed strict limitations on the way in which RNAVs can evolve, with subsequent effects on the way in which their diseases manifest in humans.
Breaking Out
However, while a virus’s polymerase is typically the major determinant of replication fidelity, it is not the only factor at play. For some time, the Nidovirales order of RNAVs were an exception to the Eigen trap, with the Coronaviridae family including viral genomes as large as 32kb — far greater than what should be feasible [12]. The paradox led to some speculation around how the nidoviruses had achieved this, yet, given that few such viruses infected humans, research was largely shelved in favor of more important projects.
However, in 2002, the highly fatal Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) emerged in Southeast Asia, propelling coronaviruses to the height of clinical relevance [13]. To the delight of many, subsequent research showed that SARS-CoV, and many other nidoviruses encoded non-structural proteins (nsp) with 3-5′ exonuclease activity [14]. By associating with viral RdRps in vivo, these exonucleases were shown to confer proof-reading activity that significantly improved replication fidelity and facilited the large size and complexity of their genomes. Therefore, while the coronaviruses still form quasispecies, their populations are significantly less diverse than other RNA viruses.
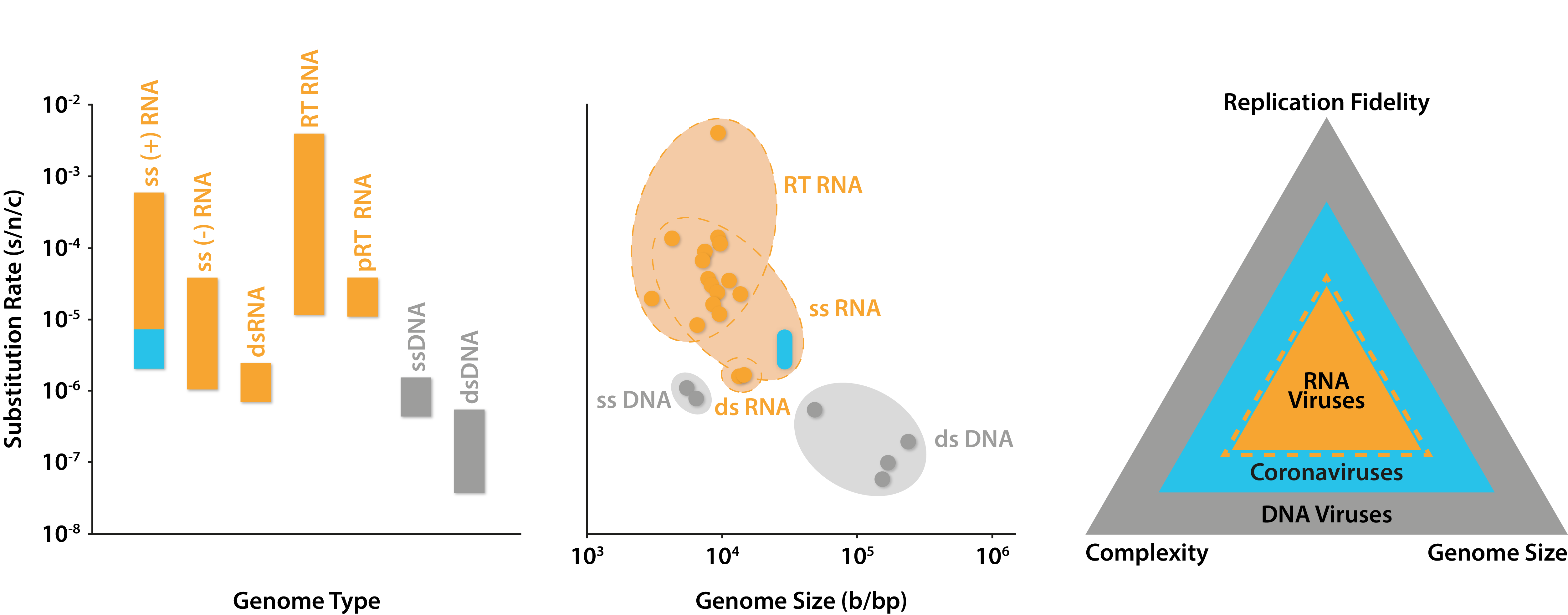
Rates of substitution across classes of human viruses. Left: substitutions per nucleotide per cell infection of different Baltimore classes with blue area indicating predicted range of coronavirus mutation rates; Middle: substitutions per nucleotide per cell infection against genome size, with blue area indicating predicted range of coronavirus mutation rates; Right: depiction of positive relationships between the size of viral genomes and their complexity and replication fidelity. Graphs adapted from Sanjuán and Domingo-Calap (2016), data from Liu and colleagues (2020) and The Royal Society (2020).
Since emerging in 2019, SARS-CoV-2 has proliferated across much of the world, causing substantial morbidity, mortality, and economic disruption. Like its coronavirus predecessors, SARS-CoV-2 encodes a proofreading 3-5′ exonuclease (nsp14) that improves polymerase (nsp12) fidelity [15]. For many, this was a positive finding, as the degree to which the novel virus could mutate appeared limited, and would have minimal impact on the ongoing development of countermeasures. Yet, over the course of a global pandemic that has infected over 75 million people, SARS-CoV-2 has had plenty of opportunity to explore new mutations.
Analyses of genomic data since summer 2020, has shown that various point mutations have occurred, with a substantial amount in the gene encoding the Spike protein [16]. Given that many countermeasures have been developed using the Spike sequence of the original Wuhan reference strain, there has been ongoing concern about their continued efficacy, as was the case for the previous coronavirus epidemics [17, 18, 19]. Furthermore, the unequal geographic and temporal distribution of vaccines and therapeutics, once available, have raised concern about the acceleration of selection pressures creating vaccine-resistance.
D614G
First identified in China in the weeks that followed SARS-CoV-2’s initial identification (then nCoV-2019), a variant with an A-to-G point mutation at position 23,403 within the Spike gene was identified that resulted in an aspartate (D) to glycine (G) mutation at residue 614 [21]. At the time, D614G was just another product of natural genetic drift and drew little attention from the scientific community. However, the G614 mutant seeded the initial outbreak in Europe shortly after, before reaching a near-global consensus [22]. Epidemiological data suggested that G614 was transmitting faster D614, leading many to speculate that the mutation had conferred enhanced infectivity [23]. D614G soon became a contentious point of debate in the scientific community, with a flurry of infectivity studies aiming to corroborate the epidemiological findings.
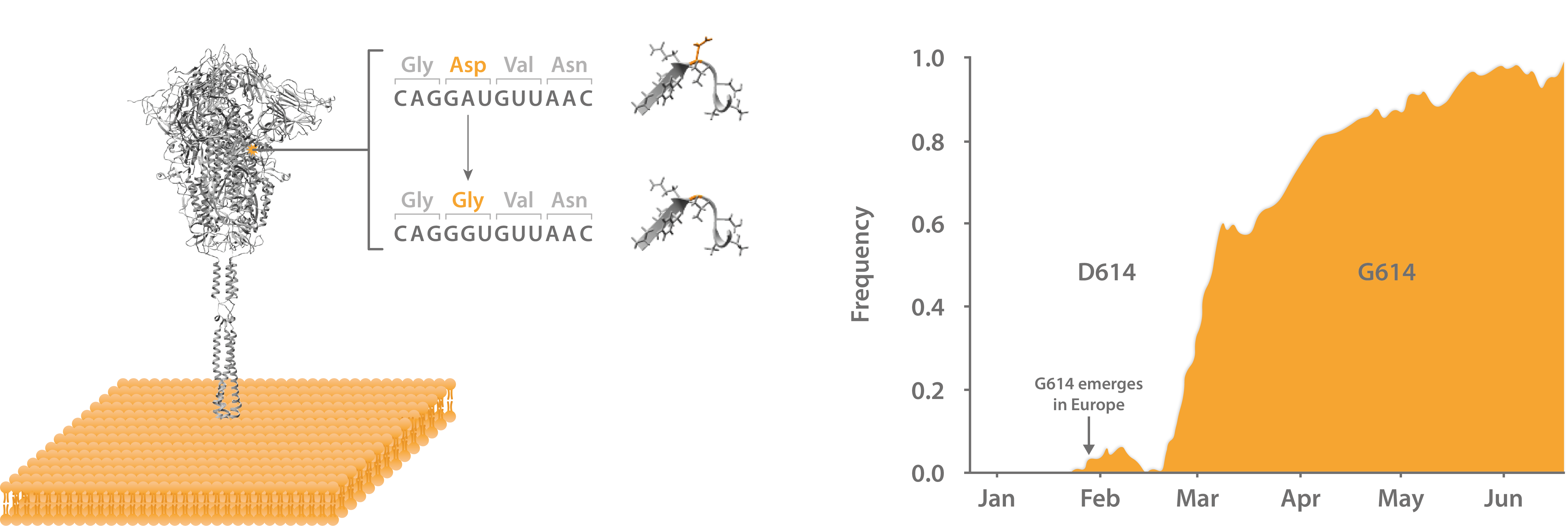
Left: Structure of SARS-CoV-2 Spike glycoprotein with D614G mutation at S1-S2 interface. Right: Frequency of global D614G incidence from isolates from Jan-Jun 2020.
Multiple infectivity studies showed that the G614 mutation conferred improved transduction efficiency, infectivity, viral loads and Spike-virion incorporation in vitro [24, 25]. Looking for a potential mechanism, structural studies identified D614G to be at the junction of Spike’s S1 and S2 subunits, where the aspartate > glycine mutation eliminated a salt bridge interaction [26, 27]. Computational modelling then showed that this could confer greater flexibility in the receptor-binding domain (RBD), so that it more frequently adopts the “open” structure required to for ACE2 binding [24].
While the evidence appeared to be building in favour of the greater infectivity hypothesis, correlating in vitro and in silico data with clinical phenotypes is notoriously fraught with pitfalls. Furthermore, some suggested that the mutant had arisen by a founder effect that had obscured the appearance of selection pressures. Host and environmental factors could then have permitted sustained clusters of infection that propagated the mutation to dominance [28].
However, corroborating the earlier findings, in vivo models largely confirmed the enhanced infectivity hypothesis. Multiple animal models studies showed G614 to yield higher viral titres [29], and enhanced droplet transmission in the upper respiratory tract [20]. Genomic data from COVID-19 patients also showed G614 to be associated with higher viral loads [24] and emerging epidemiological data suggested that a founder effect alone would be insufficient to explain D614G’s success [31].
However, while it now appears that D614G does indeed confer greater infectivity, the clinical significance of this finding is not as substantial as it might seem. Because the mutation primes Spike’s RBD to more frequently adopt an open structure, the variant has shown to be more susceptible to RBD-specific neutralising antibodies [32]. Moreover, as the mutation is not present within the RBD or N-terminal domain, to which most neutralising antibodies bind, it’s impact on antibody specificity is likely minimal. Therefore, D614G appears to have opted for infectivity in place of pathogenicity, and will likely have a negligible impact on vaccine or therapeutic efficacy [33].
Indeed, the early attention lavished on G614 may obscure a bigger question: With SARS-CoV-2 having spread to tens of millions worldwide, why aren’t more mutations that affect its behavior emerging? Clearly, the 3-5′ exonuclease has played a significant role in limiting mutations and has been highly advantageous to the development of countermeasures. However, with the continuous emergence of novel mutants over recent months, careful ongoing attention to disease surveillance will be required.
Spike Mutants from The Native Antigen Company
To support SARS-CoV-2 research, The Native Antigen Company offers a range of mutant Spike S1 glycoproteins, based on commonly identified sequences. These mutants have been produced from The Native Antigen Company’s proprietary mammalian expression system to ensure proper full folding and full glycosylation.
We are continually developing a range of mutant Spike proteins as new variants emerge. If you have a specific request, please get in touch at NAC.Contact@LGCGroup.com and we may be able to develop something for you.
References
1. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5075021/
2. https://thenativeantigencompany.com/the-yearly-race-for-an-influenza-vaccine/
3. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2660912/
4. https://www.nature.com/articles/s41564-020-0690-4
5. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5221455/
6. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000003
7. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4826558/
8. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC111862/
9. https://www.nature.com/articles/35085569
10. https://www.nature.com/articles/nature04388
11. https://pubmed.ncbi.nlm.nih.gov/4942363/
12. https://www.sciencedirect.com/science/article/abs/pii/S0966842X16301330
13. https://www.who.int/csr/don/2003_03_27b/en/
14. https://www.pnas.org/content/103/13/5108
15. https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-020-02344-6
16. https://www.biorxiv.org/content/10.1101/2020.06.25.170688v2
17. https://www.nature.com/articles/s41541-020-00246-8
18. https://pubmed.ncbi.nlm.nih.gov/18989460/
19. https://pubmed.ncbi.nlm.nih.gov/24778221/
20. https://www.pnas.org/content/117/40/24614
21. https://www.nature.com/articles/s41598-020-70827-z
22. https://www.biorxiv.org/content/10.1101/2020.07.04.187757v2
23. https://linkinghub.elsevier.com/retrieve/pii/S0092867420308205
24. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7310631/
25. https://www.biorxiv.org/content/10.1101/2020.06.14.151357v1.abstract
26. https://www.medrxiv.org/content/10.1101/2020.07.31.20166082v2
27. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7332439/
28. https://www.pnas.org/content/117/38/23652
29. https://www.nature.com/articles/s41586-020-2895-3
30. https://doi.org/10.1101/2020.09.28.317685
31. https://www.sciencedirect.com/science/article/pii/S0006291X2032074X
32. https://science.sciencemag.org/content/370/6523/1464