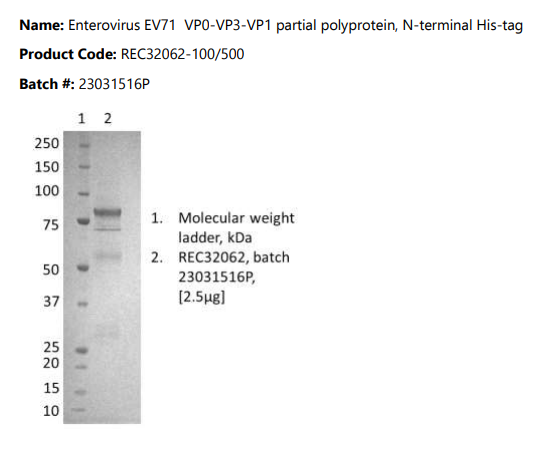
Enterovirus EV71 VP0-VP3-VP1 partial polyprotein
Enterovirus EV71 VP0-VP3-VP1 partial polyprotein, N-terminal His-tag, purified by immobilized metal affinity chromatography, followed by ion exchange chromatography and dialysis.
PRODUCT DETAILS – Enterovirus EV71 VP0-VP3-VP1 partial polyprotein
- Enterovirus EV71 VP0-VP3-VP1 partial polyprotein, EV71/Homo sapiens/KHM/349/2012 strain.
- Expressed and purified from HEK293 cells with N-terminal His-tag Native Antigen.
- NCBI Accession number: AJD77364.1
- Purified by immobilized metal affinity chromatography, followed by ion exchange chromatography and dialysis.
- Presented as liquid in 50 mM sodium phosphate pH 7.5, 100 mM NaCl buffer.
BACKGROUND
Group B coxsackieviruses (CVBs) are etiologic agents of a number of human diseases that range in severity from asymptomatic to lethal infections. They are small, single-stranded positive RNA icosahedral viruses that belong to the enterovirus genus of the picornavirus family. The coxsackievirus B3 strain Nancy (CB3) genome is 7396 nucleotides long, and encodes a 2185 amino acid long polyprotein. With only one open reading frame, CVB genome encodes a large polyprotein, which is cleaved into the mature viral proteins by viral proteinases 2A and 3C (Lindberg et al., 1987, Sean and Semler, 2008). CVB capsid is composed of four structural proteins, VP1, VP2, VP3, and VP4. The VP1 protein, which locates on the surface of the capsid, is the main neutralizing antigen of the virus (Muckelbauer and Rossmann, 1997). It exhibits the same gene organization as other enterovirus genomes (Lindberg et al., 1987).
Capsid protein VP1 forms an icosahedral capsid of pseudo T=3 symmetry with capsid proteins VP2 and VP3. The capsid is 300 Angstroms in diameter, composed of 60 copies of each capsid protein and enclosing the viral positive strand RNA genome (Halim and Ramsingh, 2000). The β-sandwich structure of the viral capsid proteins VP1, VP2 and VP3 is conserved between CVB3 and other picornaviruses and structural differences between CVB3 and other enteroviruses and rhinoviruses are located primarily on the viral surface (Muckelbauer et al., 1995). VP1 mainly forms the vertices of the capsid and interacts with host CD55 and CXADR to provide virion attachment to target host cells. This attachment induces virion internalization. Tyrosine kinases are probably involved in the entry process. After binding to its receptor, the capsid undergoes conformational changes. VP1 N-terminus (that contains an amphipathic alpha-helix) and capsid protein VP4 are externalized. Together, they shape a pore in the host membrane through which viral genome is translocated to the host cell cytoplasm. After the genome has been released, the channel shrinks (Halim and Ramsingh, 2000). VP1 is not only a structural unit of the capsid, but also involved in viral pathogenesis. A single amino acid mutation in the VP1 protein can significantly change antigenicity, where a virulent variant becomes more antigenic than an avirulent variant (Halim and Ramsingh, 2000).
REFERENCES
- Halim and Ramsingh (2000). A Point Mutation in VP1 of Coxsackievirus B4 Alters Antigenicity. Virology. Volume 269, Issue 1, Pages 86-94.
- Lindberg et al. (1987). Genome of coxsackievirus B3. Virology. Volume 156, Issue 1, Pages 50-63.
- Muckelbauer et al. (1995). The structure of coxsackievirus B3 at 3.5 å resolution. Structure. Volume 3, Issue 7, July 1995, Pages 653-667.
- Wang et al. (2012). A functional nuclear localization sequence in the VP1 capsid protein of coxsackievirus B3. Virology. Volume 433, Issue 2, Pages 513-521.