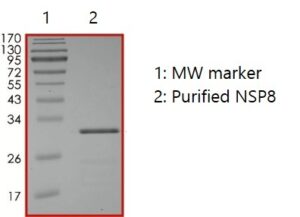
SDS-PAGE: Coomassie-stained reducing SDS-PAGE showing purified NSP8.
SARS-CoV-2 NSP8 Protein, His-Tag (E. coli)
Price range: $531.83 through $1,019.50 excl. VAT
SARS-CoV-2 NSP8 protein, His-tag is a recombinant full-length viral protein expressed in E. coli. SARS-CoV-2, previously known as the 2019 Novel Coronavirus (2019-nCoV), causes the pandemic COVID-19 disease.
SARS-COV-2 NSP8, HIS-TAG (E. COLI)
SARS-CoV-2 NSP8 protein, His-tag is a recombinant full-length viral protein expressed in E. coli. SARS-CoV-2, previously known as the 2019 Novel Coronavirus (2019-nCoV), causes the pandemic COVID-19 disease.
PRODUCT DETAILS – SARS-COV-2 NSP8, HIS-TAG (E. COLI)
- SARS-CoV-2 NSP8 protein, His-tag.
- Expressed in E. coli cells with an C-terminal His-tag and >90% purity.
- Presented as liquid in 520 mM Tris, pH 7.5, 300mM NaCl, 200mM imidazole, 1 mM DTT, 10% glycerol.
BACKGROUND
In December 2019 a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), formerly known as the 2019 novel coronavirus (2019-nCoV) was identified in Wuhan, China, causing a world-wide pandemic (Wu et al., 2020). Three coronaviruses, SARS-CoV, MERS-CoV, and SARS-CoV-2 have been identified as being a highly pathogenic for humans, and there is currently no effective antiviral treatment. Therefore, studies are focused on rapid development of vaccines and antiviral drugs to prevent and treat coronavirus infection. There are several potential strategies to pharmacologically fight against the disease (COVID-19), including vaccines, monoclonal antibodies, oligonucleotide-based therapies, peptides, interferon therapies, and small-molecule drugs (Dömling & Gao, 2020).
Coronavirus RNA genome encodes a complex replication machinery consisting of 16 viral non-structural proteins (NSPs). NSP7 and NSP8 stabilize regions of NSP12 (RNA-dependent RNA polymerase; RdRp) involved in RNA binding and are essential for a highly active NSP12 polymerase complex. NSP7 has been suggested to confer RNA-binding properties to the trimeric polymerase complex (Subissi et al., 2014). NSP8 has been shown to act as an oligo(U)-templated polyadenylyltransferase and a robust (mono/oligo) adenylate transferase (Tvarogova et al., 2019). The NSP7/NSP8/NSP12 polymerase complex associates with NSP14, which is required to safeguard coronavirus replication fidelity (Subissi et al., 2014). Specific mutations in NSP7 and NSP8 may have a role in stabilization of the replication/transcription complex (Reshamwala et al., 2021). Amino acid substitutions in RdRp (P323L) have been shown to cause conformational changes near the NSP8 binding site that may affect virus replication and transcription (Alosaimi et al., 2021).
REFERENCES
- Alosaimi B, Naeem A, Alghoribi MF, Okdah L, Hamed ME, AlYami AS, Alotaibi A, Enani M. Structural Mapping of Mutations in Spike, RdRp and Orf3a Genes of SARS-CoV-2 in Influenza Like Illness (ILI) Patients. Viruses. 2021 Jan 19;13(1):136.
- Dömling A, Gao L. Chemistry and Biology of SARS-CoV-2. Chem. 2020;6(6):1283-1295.
- Reshamwala SMS, Likhite V, Degani MS, Deb SS, Noronha SB. Mutations in SARS-CoV-2 nsp7 and nsp8 proteins and their predicted impact on replication/transcription complex structure. J Med Virol. 2021 Jan 12.
- Subissi L, et al: One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. PNAS. U.S.A. 2014, 111: E3900–E3909.
- Tvarogova J, et al: Identification and characterization of a human coronavirus 229E nonstructural protein 8-associated RNA 3′-terminal adenylyltransferase activity. Virol. 2019, 368: 779-782.
- Wu F. et al: A new coronavirus associated with human respiratory disease in China. Nature 579: 265-269, 2020.