The Native Antigen Company is now offering recombinant S1 and S2 glycoproteins for SARS-CoV-2 (Covid-19) in response to urgent demand. These reagents are suitable for use in basic research and the development of diagnostics and vaccines.
These antigens have been produced using The Native Antigen Company’s proprietary VirtuE (HEK293) expression system, to ensure full glycosylation and proper folding.
The Coronavirus Spike Proteins
The ability of Coronaviruses to gain entry to cells and establish infection is mediated by the interactions of its Spike glycoprotein with human cell surface receptors. The coronavirus Spike proteins are large type I transmembrane protein trimers that protrude from the surface of coronavirus virions. Each Spike protein comprises three segments: A large ectodomain (comprising S1 and S2), a transmembrane anchor, and a short intracellular tail [1]. The S1 subunit of the ectodomain mediates binding of the virion to host cell-surface receptors through its receptor-binding domain (RBD). The S2 subunit fuses with both host and viral membranes, by undergoing dramatic structural changes [2].
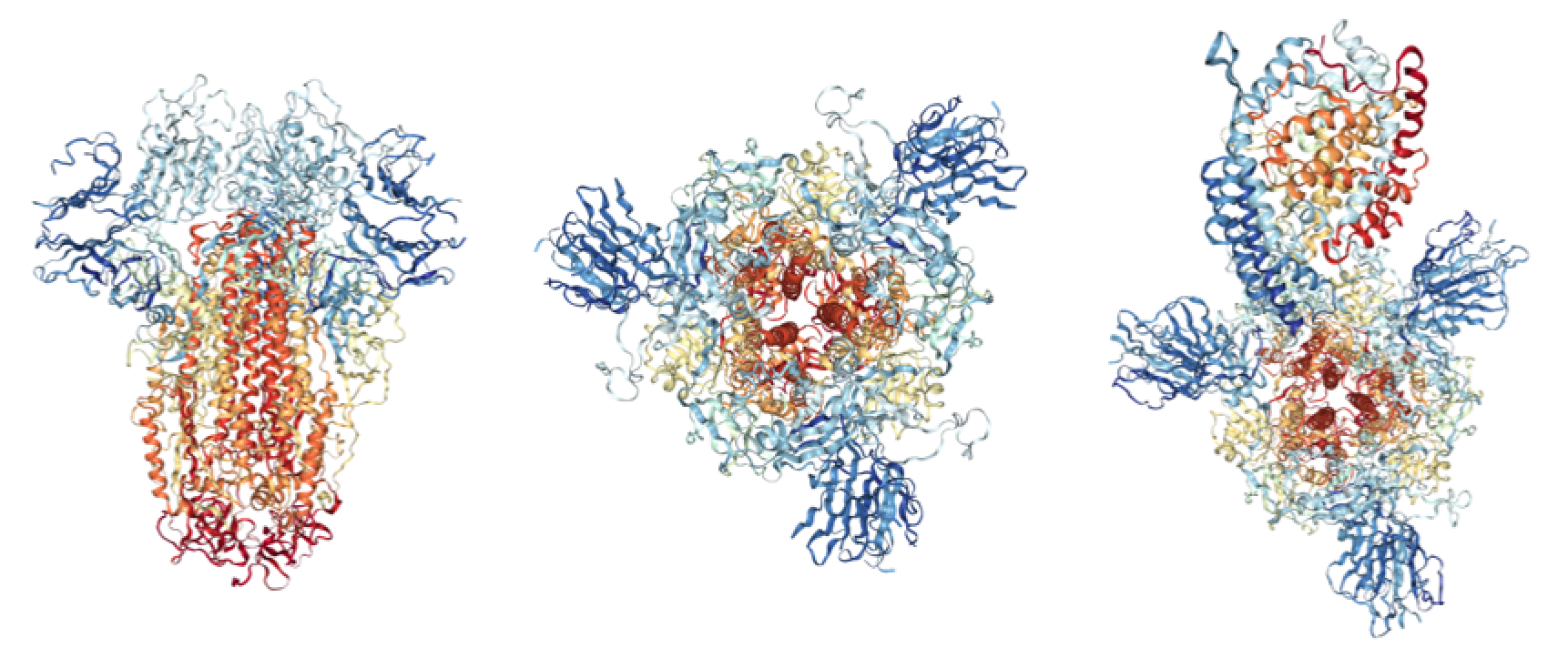
Crystal structures of SARS-CoV Spike protein. Left: Spike protein side-on view (RCSB: 5XLR); Middle: Spike protein top-down view; Right: Spike protein bound to ACE2 receptor top-down view (RCSB: 6ACJ).
Structural Studies on SARS-CoV-2’s Spike Protein
As the Spike’s receptor binding domain is the primary determinant of a coronavirus’s ability to infect a host species and its tropisms, it has been the focus of several early studies.
Alignment of the SARS-CoV-2’s genome shows that its closest human-infecting relative is the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), with a sequence identity of approximately 79% [3]. Identity between the spike proteins is 76.47%, with slightly lower sequence identity between the RBDs (73%) [4]. Within SARS-CoV-2’s 193 residue RBD, a large proportion of mutations are highly accumulated at the C-terminal region that includes the receptor binding motif [5].
Modelling by Xu and colleagues suggests that these differences do not significantly affect Spike’s binding affinity for ACE2 as the Spikes share an almost identical 3D structure in the RBD, despite sequence divergence, which has been corroborated by modelling and infectivity studies by various groups [5, 6, 7]. Wan and colleagues showed that several critical residues in SARS-CoV-2’s receptor-binding motif (RBM) are compatible with, but not ideal for binding of human ACE2 (e.g. Asn501). Therefore, while SARS-CoV-2’s is capable of human-to-human transmission, it appears to bind suboptimally with ACE2 when compared with the 2002 SARS-CoV strain [5]. However, considering that this region is highly looped, variations are likely to cause structural rearrangements that allow for novel and possibly even stronger interactions with ACE2 [8]. Indeed, it has been suggested that a single N501T mutation in SARS-CoV-2’s Spike protein could significantly enhance its binding affinity for ACE2 [5], and considering that the SARS’s Spike protein mutated over the course of the 2002-2004 epidemic to better bind human cell-surface receptors, this is not an unlikely scenario.
Pradhan and colleagues described 4 unusual SARS-CoV-2 Spike protein insertion sequences that are discontinuous in the amino acid sequence but are thought to converge at the RBD. Alignments showed that they are not present in any other coronavirus strains but show identity/similarity with sequences in HIV-1 gp120 and Gag, the former of which is also a cellular receptor recognition protein [10]. The authors speculate that these insertions confer additional flexibility to the RBD by forming a hydrophilic loop that mediates virus-host interaction with greater infectivity. While the authors do not explicitly state that these sequences have been gained from HIV-1, they note that their presence is “unlikely to be fortuitous in nature”. It is worth noting that this paper has led some to speculate that SARS-CoV-2 has been deliberately engineered with additional HIV-1 sequences. However, considering that these inserts appear in hypervariable regions of the protein and are as short as 6 residues in length, it is very probable that they arose naturally.
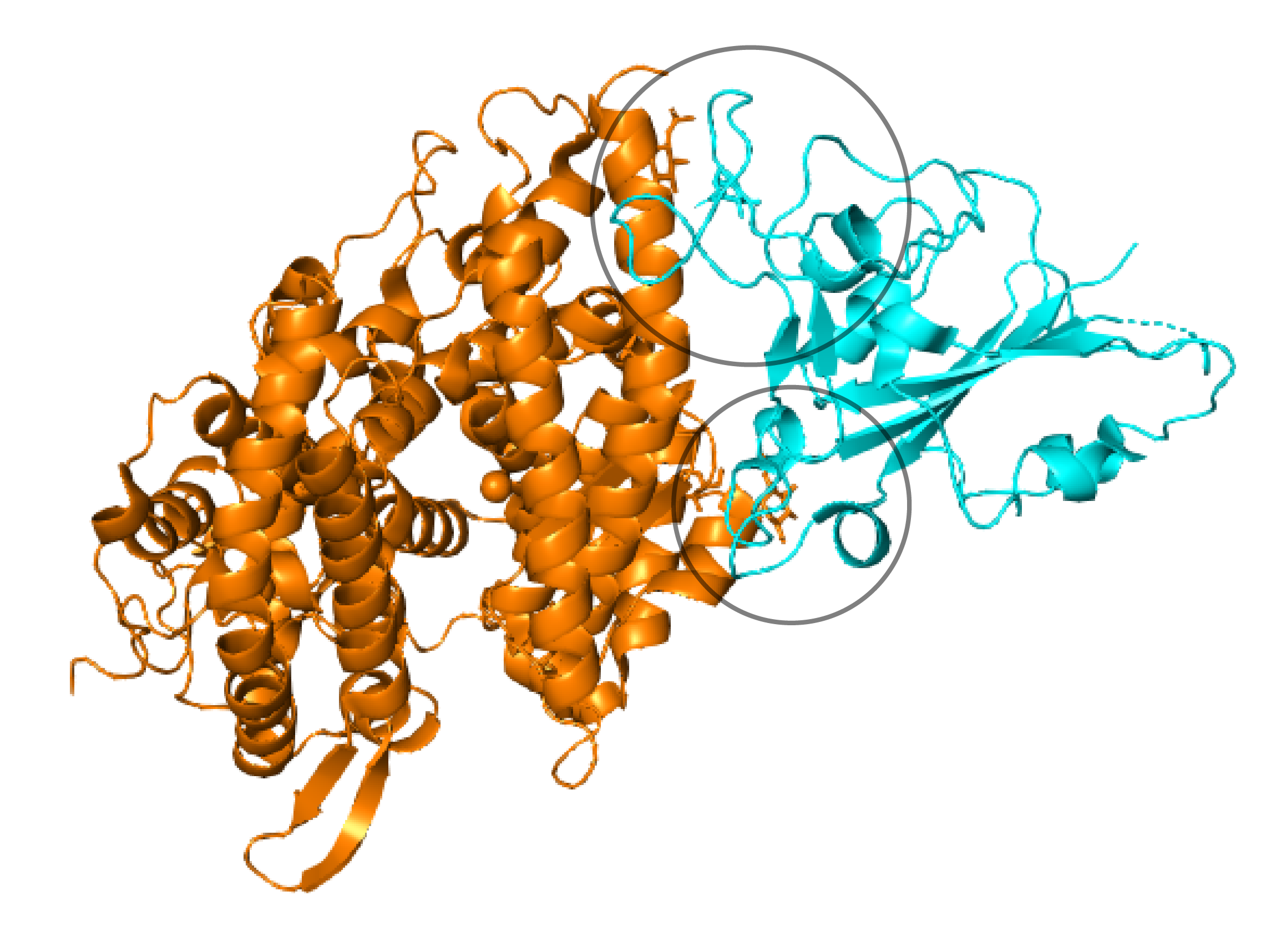
Crystal structure of Spike RBD (cyan), bound to ACE2 cell surface receptor (orange). Variable loop regions that include insertion sequences in the 2019 SARS-CoV-2 are circled (RCSB: 3D0I).
Immune Responses to Spike Proteins
The Spike protein is the central target of therapeutic and vaccine development for the Novel Coronavirus [1]. However, in order to develop effective countermeasures that can prevent and treat infection, characterisation of SARS-CoV-2’s immune responses and correlates of protection will be necessary.
Coronavirus Spike proteins are known to play a key role in eliciting potent neutralising-antibody and T-cell responses [11]. The SARS Spike protein in particular has been found to elicit a range of neutralising antibodies through its major immunodominant S1 epitope (residues 441 to 700) [12]. Interestingly, Tian and colleagues showed that many of the 40 most potent SARS-CoV-specific neutralising antibodies that target its RBD failed to bind SARS-CoV-2’s spike protein. This suggests that structural differences in the RBD of SARS-CoV and SARS-CoV-2 have a significant impact on the cross-reactivity of neutralising antibodies, and suggests that novel monoclonals will need to be identified for therapeutic and vaccine development. In their study, Tian and colleagues did manage to identify that monoclonal antibody CR3022, bound SARS-CoV-2’s Spike RBD with high affinity. This is likely due to the antibody’s epitope not overlapping with the divergent ACE2 receptor binding motif. As such, CR3022 has the potential to be developed as a therapeutic candidate, alone or in combination with other neutralising antibodies, for prevention and treatment of SARS-CoV-2’s infections.
Whilst neutralising antibodies are able to prevent viral entry, the body also requires CoV-specific CD4+ T helper cells for the development of specific antibodies, as well as the recognition and killing of infected cells, particularly in the lungs of infected individuals [11]. Therefore various lines of research into cell-mediated immune responses will be also be necessary.
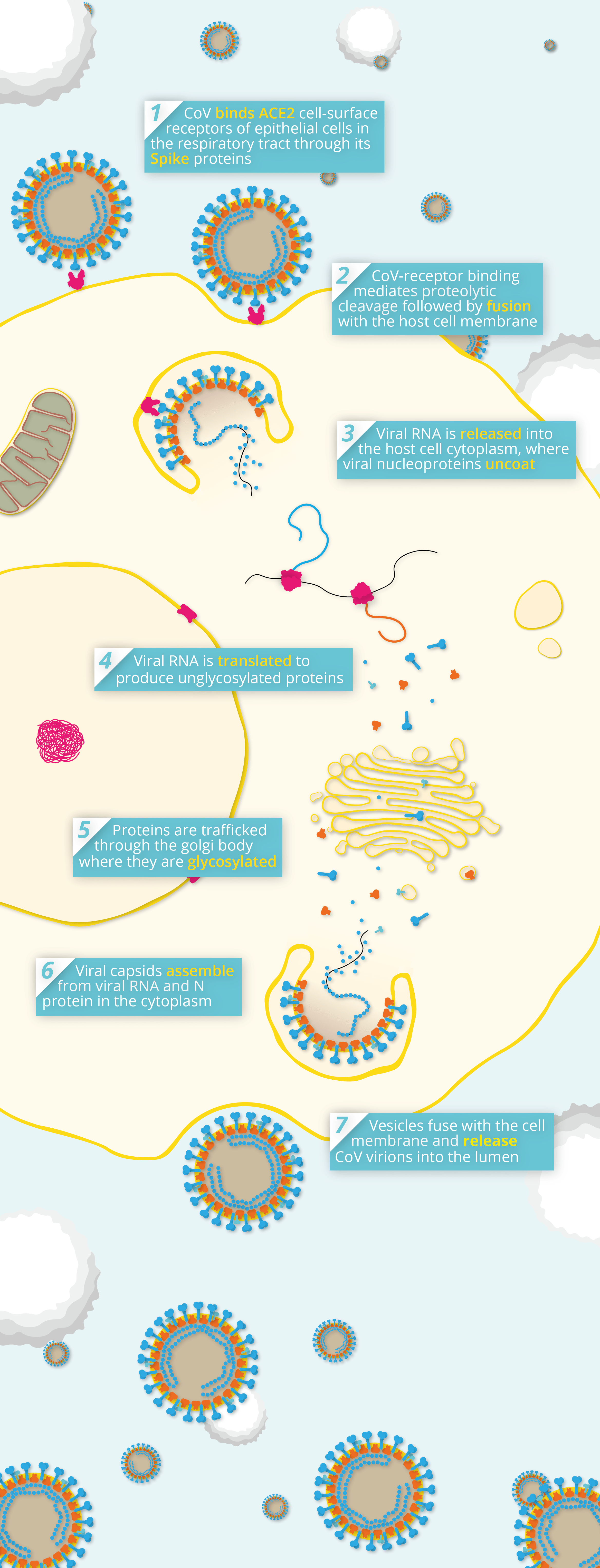
Simplified pathway of coronavirus pathogenesis.
About The Native Antigen Company
Founded in 2010, The Native Antigen Company has rapidly become one of the world’s leading suppliers of reagents for infectious disease. Over the last 10 years, the Native Antigen Company has developed rapid response manufacturing capabilities to produce reagents for emerging epidemic diseases, guided by the World Health Organisation’s R&D Blueprint, which outlines the accelerated development of vaccines and increased availability of accurate and reliable diagnostics for priority emerging infectious diseases. The Native Antigen Company’s proprietary mammalian, VirtuE expression system from which these reagents have been expressed, is able to introduce proper protein folding and post-translational modifications to recombinant proteins, which are essential for full biological and antigenic activity. The company is widely known for its expertise in this field, having used its VirtuE expression system to rapidly develop antigens for other major health threats, including the recent Zika and Dengue outbreaks.
References
1. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2750777/
2. https://jvi.asm.org/content/86/5/2856
3. https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30251-8/fulltext
4. https://link.springer.com/article/10.1007%2Fs11427-020-1637-5
5. https://jvi.asm.org/content/early/2020/01/23/JVI.00127-20.long
6. https://www.biorxiv.org/content/10.1101/2020.01.31.929042v1
7. https://www.biorxiv.org/content/10.1101/2020.01.22.914952v1
9. https://www.biorxiv.org/content/10.1101/2020.01.30.927871v1
11. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3636424/
12. https://www.biorxiv.org/content/10.1101/2020.01.28.923011v1