Following the latest immunofluoresence data for our Zika and Dengue antibodies, we’ve invited VRS to write a short blog one of their areas of expertise: The applications of immunofluorescence microscopy in studying viruses.
A lot of what we know about biology is because of the availability of antibodies. Besides being key players in diagnostics and therapeutics, these incredibly versatile and specific molecules have allowed scientists to discover where proteins are (if they are there at all!), how they interact, and (sometimes) what their surfaces look like.
Antibodies are indispensable tools in molecular studies of viral infection, as they allow the application of modern and state-of-the-art imaging technologies without having to engineer the virus. Using our expertise in microscopy and high-throughput screening, here at VRS we are working to develop new tools to better understand and treat viral infection. Good quality, specific antibodies are paramount to these studies, and The Native Antigen Company’s extensive antibody collection is allowing us to do exactly this (you can read more about our collaboration here).
Antibodies in fluorescence microscopy
In the most basic applications, antibodies recognising a viral antigen can tell us whether cells or tissues are infected. Both low-resolution microscopy and flow cytometry can be used to this end. When antibodies are unavailable, virologists can resort to following the consequence of infection, which is usually cell death. The virus can be left to spread freely across a cell culture and cytopathic effect (CPE) can be measured either by eye or using dyes that distinguish live from dead cells. The concentration of virus where 50% of CPE is observed is generally presented as an indication of infectivity. Alternatively, the virus can be forced to spread exclusively cell-to-cell (rather than freely in the media) by covering the culture with a solid or semisolid overlay, which allows to visualise infection in neat and defined plaques.
These methods work very well for viruses that cause CPE and do so fairly rapidly. For all the others, antibodies are an invaluable tool for determining percentages of viral infection, or to stain for plaques when the virus would take too long to cause a proper “hole” of dead cells in the monolayer. This is the case with dengue virus: immunostaining of infected foci allows quantification of infectious virus in 3 days rather than >7 days. Although more expensive than traditional CPE-based methods, immunofluorescence has the advantage of providing clearer and more easily quantifiable results: many factors can cause holes in a monolayer of cells, but good antibodies guarantee specificity.
Time courses also determine how and at what speed a virus spreads: how long does it take to go from one infected cell to many? And does it spread by releasing lots of new virions in the extracellular space, infecting cells further away, or by remaining tightly associated to its first target and quietly spreading to the neighbours? Immunofluorescence staining and microscopy allow us to distinguish between these different patterns.
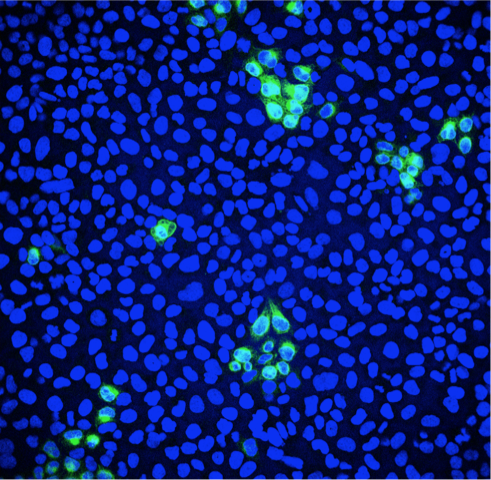
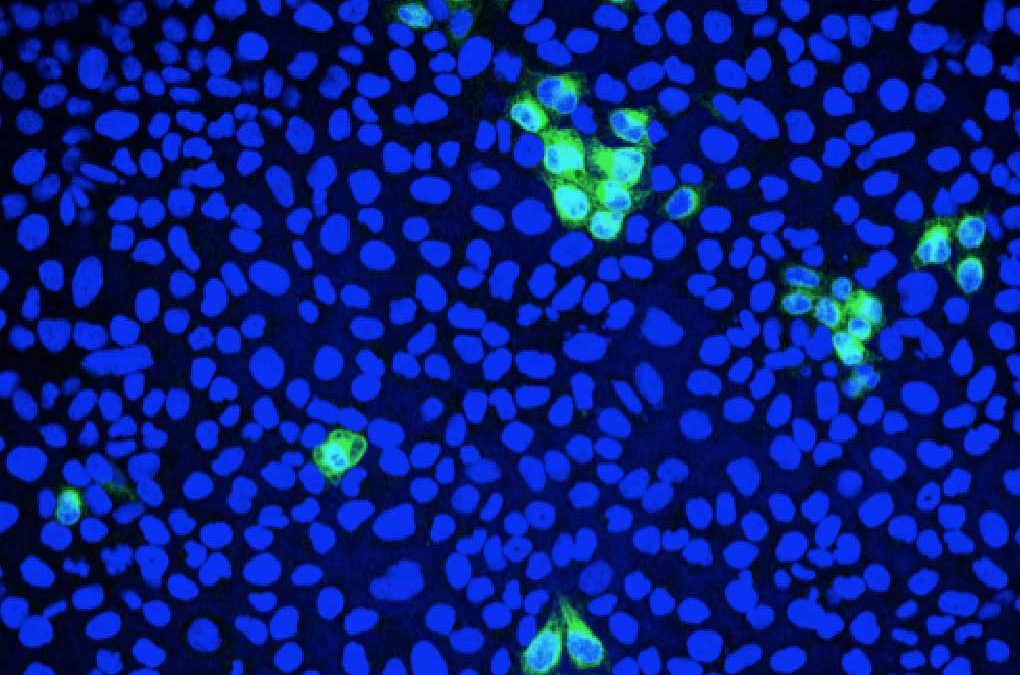
Fluorescence microscopy can be used to quantify the percentage of infected cells. Green: Zika virus infected cells; Blue: cell nuclei.
Antibodies in high-resolution microscopy
Quantification is extremely important, but high-resolution imaging is probably the most exciting application of antibodies to the field of virology. This is when the virus comes alive and its intimate interaction with the host cells is exposed. Staining for viral antigens, for instance, allows us to determine where these antigens are, which is often a good indication of where the virus replicates. Many flavivirus proteins can be detected in the endoplasmic reticulum, where the virus replicates. Alphaviruses tend to accumulate in distinct clusters in the cytoplasm or near the cell membrane. Influenza quickly goes into the nucleus. By co-staining for cellular proteins or organelles, it is also possible to look at the co-localisation between virus and cellular compartments, and to see whether and how the virus manipulates and reorganises the cellular environment to replicate as efficiently as possible. A good antibody against a viral surface protein is an even more useful tool: under high-resolution, it is possible to follow the virus while it binds and enters the cell, thus gaining knowledge of the early stage of viral infection. Unfortunately, antibody staining of intracellular antigens is not compatible with live imaging, which is only made possible by fluorescently tagged viruses. While tagging of large DNA viruses is fairly straightforward, even with relatively big tags as GFP (approximately 26 KDa), this process is much more complicated for RNA viruses carrying much smaller and unstable genomes. For many of these viruses, antibodies are often the tool that allows us to see them in action.
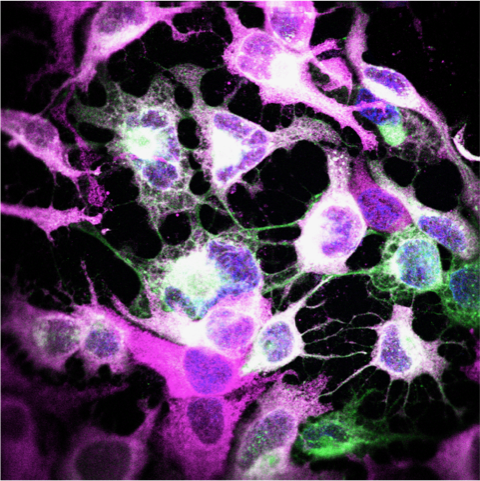
High-resolution microscopy can be used to visualise sites of viral replication. Green: Zika virus infected cells; Magenta: endoplasmic reticulum; Blue: cell nuclei.
Antibodies in high-throughput and high-content applications
High-throughput and high-content screenings are at the forefront of antiviral development. Using antibodies to visualise infection in hundreds of thousand of wells is generally not recommended (it is expensive and time consuming!), but antibodies remain an invaluable tool to re-test hit compounds against additional or more relevant viral strains after, for instance, screening by CPE or with a GFP-tagged virus. On the contrary, smaller libraries are highly amenable to immunofluorescence staining, providing the opportunity to study the virus strain of choice, as long as a good antibody is available.
Recent advances in high-throughput/high-resolution microscopy have combined the best of both worlds: the speed of high-throughput microscopy with the high-resolution of conventional imaging. These types of screening generate detailed information on the phenotype of a virus-infected cell, allowing investigators to collect a lot more information (both on the virus and the cell) upon treatment with drugs or siRNA. These studies are invaluable for our understanding of the biology of viral infection and the factors that contribute to viral replication.
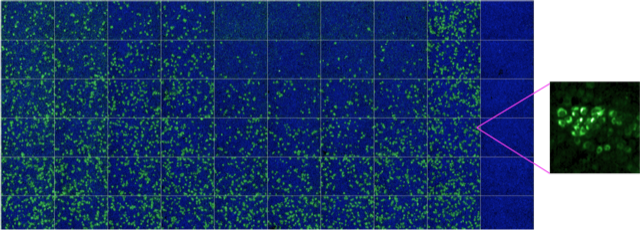
High-throughput/high-content microscopy can be used to rapidly screen thousands of compounds, or calculate dose-response curves for drugs and antibodies. Informative high-resolution images can be extrapolated to provide additional information at subcellular level. Green: Zika virus infected cells; Blue: cell nuclei.
At VRS, we make the most of the high-throughput and high-resolution facilities at University College London (MRC LMCB) to set up innovative and informative virology assays, and we collaborate with companies like The Native Antigen Company to explore the potential of new products and resources. If you want to take a closer look at your virus we can certainly help!