In the second of a three-part series on COVID-19 vaccines, we explore the potential challenges in stimulating safe vaccine responses and outline the role that diagnostics will play in guiding their development.
Rogue Responses
Antibodies play a crucial role in controlling viral infections, including sterically blocking virion-cell attachment, inducing lysis via opsonisation, and activating complement cascades [1]. Yet, while they are generally regarded as protective and beneficial mediators of the immune system, a growing body of research has suggested that not all antibodies are born equal. A serious concern for vaccine developers is the risk of antibody-dependent enhancement (ADE) – a phenomenon in which antibodies raised against a previous infection or vaccination can enhance the pathogenesis of a subsequent infection, by the same or similar virus.
First characterised as an immunological curiosity, disease-enhancing antibodies have now been implicated as critical players in the progression of many viral diseases, including dengue, HIV, influenza, RSV, Ebola and the coronavirus family [2]. The mechanism of ADE is not entirely understood and appears to vary with pathogen and patient. However, it is generally assumed that non-neutralising antibodies can mediate internalisation of a virus by Fc receptors commonly found on the surface of antigen-presenting cells (APCs) [3]. This ‘trojan horse’ strategy allows the invading virus to infect a cell, escape from its endosome, and induce altered immune mediators that impair the immune system to facilitate a more aggressive pathogenesis.
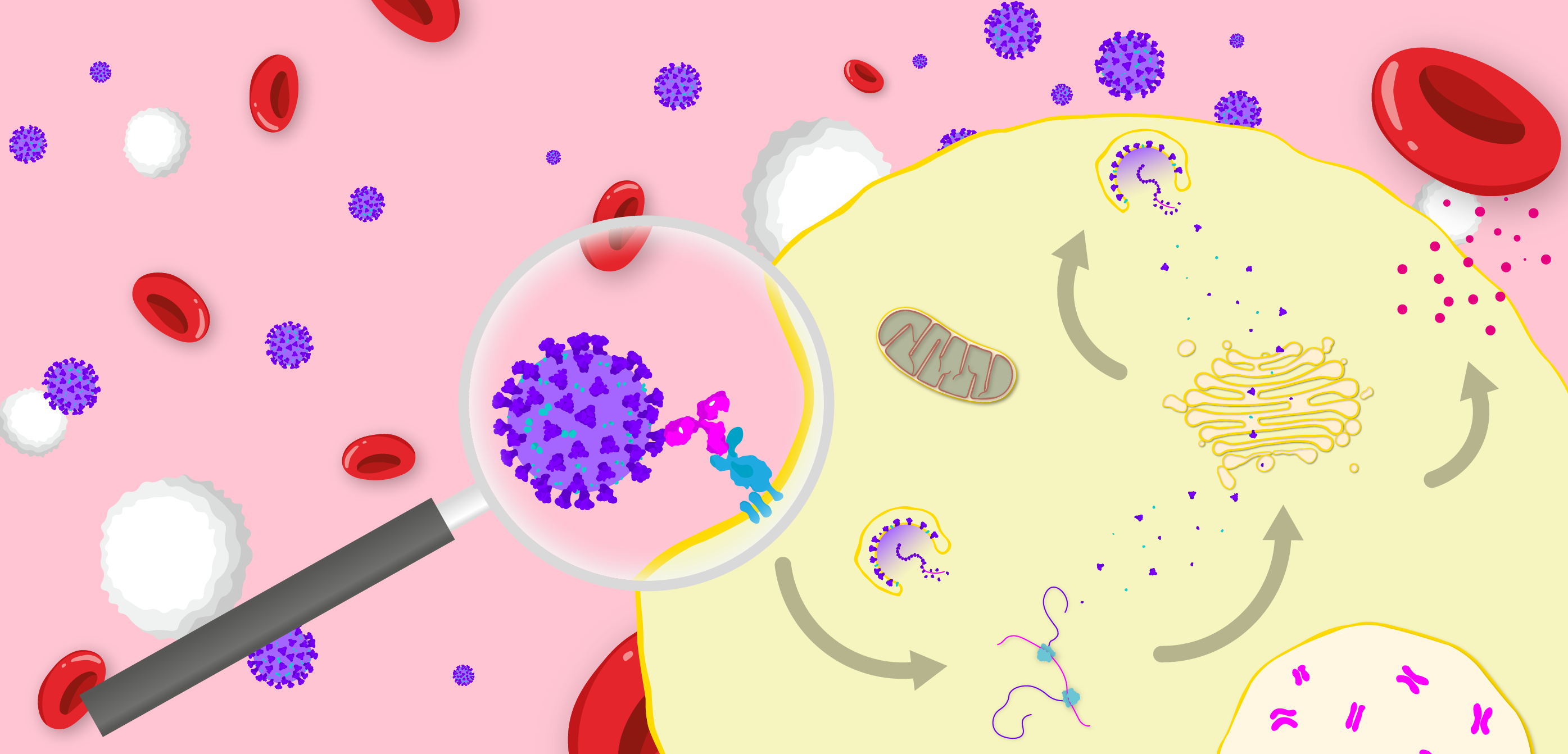
Diagram illustrating the FcγR-mediated mechanism of antibody-dependent enhancement of disease. Virus binds a non-neutralising antibody, which binds an Fcγ receptor, that in turn induces endocytosis of the viral particle. The virus then replicates within the host cell to produce progeny and stimulate the release of inflammatory cytokines.
The link between ADE and coronavirus infection harks back to the 1970s when a subunit vaccine was undergoing development for a cat coronavirus known as feline infectious peritonitis virus (FIPV). Paradoxically, vaccination increased the severity of disease in kittens and caused some deaths that quickly derailed the candidate’s development [4]. Follow-up studies showed that a combination of neutralising antibodies (nAbs) at sub-neutralising titres and non-nAbs enhanced FIPV infection of macrophages [5], releasing inflammatory and apoptotic cytokines that caused multiple organ damage [6]. Fortunately, further studies were able to mitigate some of these issues and a vaccine for FIPV is now commercially available, however its safety and efficacy is still a controversial matter of ongoing debate [7].
Fast forward to 2002 and the Severe Acute Respiratory Syndrome (SARS) coronavirus outbreak was escalating in Southeast Asia, beckoning a slew of companies to develop vaccines [8]. However, in the rush to move candidates into early clinical trials, there was an all-too-familiar sense of déjà vu. Inoculation of rhesus macaques with a Spike-encoding MVA vaccine, for example, did not protect from viral challenge and instead, elicited strong antibody responses that correlated with severe lung damage [10]. Inactivated, subunit and VLP vaccines administered to ferrets and mice then showed the same distinctive lung pathologies [11]. Interestingly, serological studies in SARS patients even appeared to mirror these findings, as the speed and degree of the antibody response was associated with rapid declines in antibody titres that correlated with lung damage and death [12, 13, 14]. Alternatively, the severity of many SARS cases peaked 3 weeks after the onset of symptoms once viral loads had begun decreasing, suggesting immunopathological mechanisms that were not driven by acute-phase infection [15].
Further investigation revealed that anti-Spike antibodies were responsible for facilitating infection of FcγRI+ (CD64), FcγRII+ (CD32) and ACE2– monocytes, macrophages and B-cells in vitro, suggesting distinct cellular pathways to canonical ACE2-mediated viral entry [15, 16, 17]. Predictably, infection of these leukocytes was implicated in the cytokine storm syndromes (CSS) and acute respiratory distress syndromes (ARDS) that led to the multiple organ failure and death, in both patients and animal models [18]. A further study by Liu and colleagues found that anti-SARS Spike IgG specifically enhanced infection of alveolar macrophages in rhesus macaques, leading to a skewed inflammatory response and lung damage that could be prevented by antibody-mediated blockade of FcγRs.
Yet SARS disappeared as fast as it had emerged. The lack of funding by 2008 caused research to dry-up and interest inevitably waned [20]. As such, no vaccines made it past Phase I trials and the potential effects of ADE could not be investigated in large human cohort studies. A decade later, MERS emerged in the Middle East, but it was a similar story: vaccinated animals showed the same characteristic lung pathologies when exposed to live virus [21], with in vitro studies implicating the potential involvement of enhancing antibody responses [22].
The Emerging Immunology of COVID-19
Fast forward a decade and the world now finds itself in the grip of the most prolific coronavirus to-date. At the time of writing, worldwide cases of Coronavirus Disease 2019 (COVID-19), caused by SARS-CoV-2 exceed 3,600,000 and deaths sit at just over 250,000 [23], though both likely a gross underestimate of the true toll. The pandemic has pushed healthcare systems to their limits, paralysing the global economy and sparking one of the biggest technology races in history.
Like its predecessors, COVID-19 manifests with a broad range of clinical symptoms. The majority of cases are thought to be asymptomatic or present with mild flu-like symptoms, while around 10% experience a severe pneumonia that requires hospitalisation [24]. Infection and destruction of cells in the lungs triggers a local immune response that recruits macrophages and monocytes, which in turn release cytokines and prime the adaptive T- and B-cell responses. In most cases, this process is capable of resolving the infection within a couple of weeks. However, in those less fortunate, a dysfunctional inflammatory response attracts an abundance of immune cells from the blood, which further potentiate cytokine release [25, 26]. If unchecked, this can cause lung damage and multiple organ failure of the cardiac, hepatic and renal systems, typical of the ARDS and CSS-like pathologies of other betacoronavirus infections [27, 28, 29, 30].
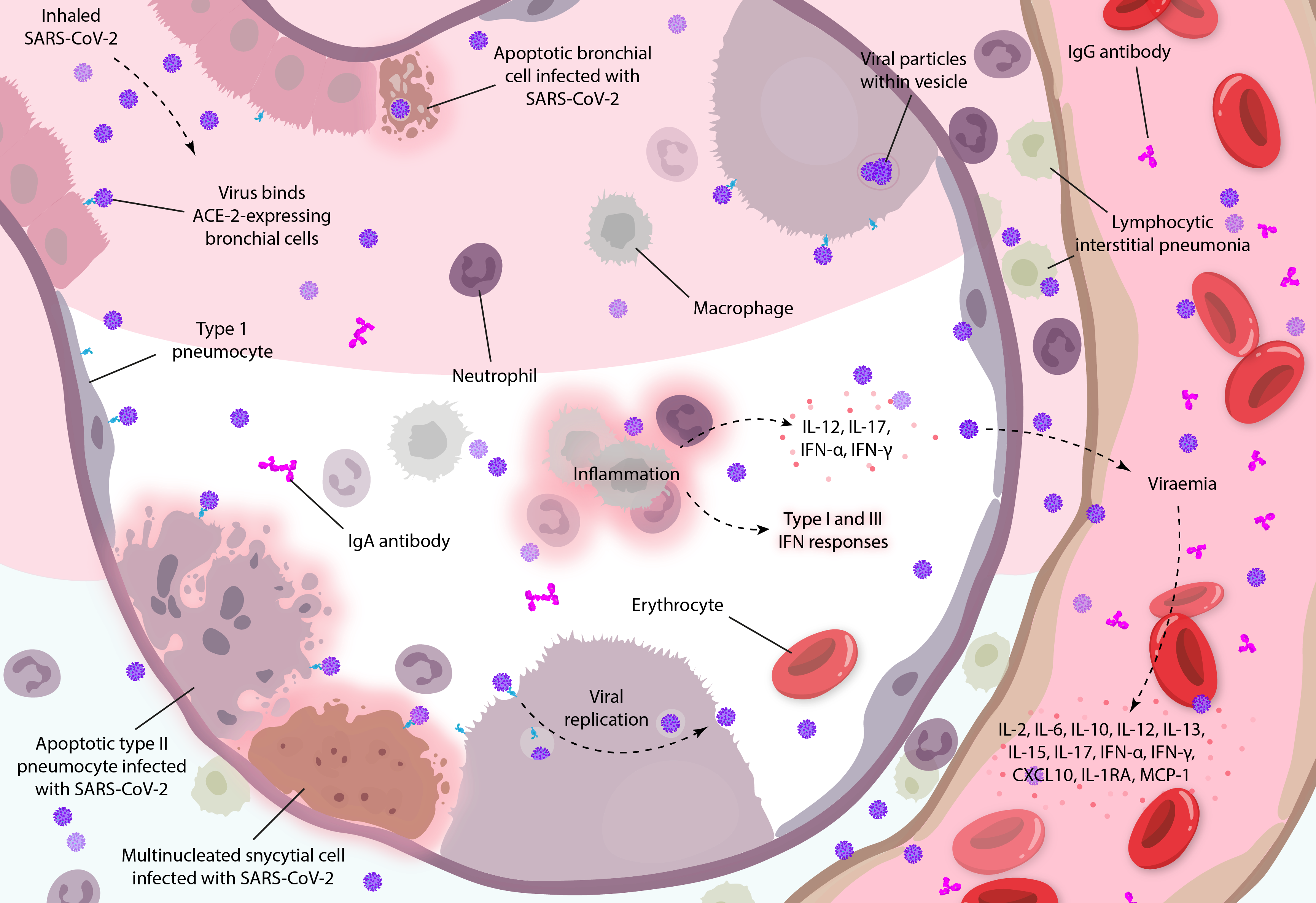
Diagram illustrating how coronavirus infections can mediate systemic inflammatory responses in the lungs, based on clinical observations and studies in vitro [19, 31]. Diagram adapted from Arabi and colleagues.
The serology of SARS-CoV-2 also bears similarities to SARS and MERS-CoV. The nature and degree of antibody responses in those with COVID-19 strongly correlates with age, elevated levels of pro-inflammatory cytokines and worsening disease prognosis [33], and COVID-19 patients that develop early and potent antibody responses appear to be at higher risk of rapidly declining antibody titres, associated with the most severe forms of disease [12, 29, 34, 35]. Conversely, children appear to respond with reduced levels of IgG at lower affinities, that negatively correlate with severity of infection [36, 37]. Similar to observations in SARS patients, many severe COVID-19 cases also tend to manifest late in the time-course of disease in a biphasic manner that corresponds to decreasing viral loads and increased IgG titres [38], suggesting immunopathological mechanisms that aren’t driven by acute infection.
The reason for the subsequent loss of antibodies in those that rapidly seroconvert is unknown. However, associated indicators of immune exhaustion could be due to a loss of leukocytes (namely B-cells), which have shown to be significantly lower in these patients [39]. Likewise, post-mortem analyses in those that have succumbed to COVID-19 suggest that infection and apoptosis of resident macrophages results in the prolific IL-6 release that contributes to damage of the lungs, heart, spleen, lymph nodes [40]. While such inflammatory responses are typically kept in check by Th2 cells, around one third of COVID-19 patients show evidence of depleted and exhausted T-cells, indicating an inability to control systemic inflammation [41, 42, 43], associated with increased levels of IL-6, IL-10 and TNF-α [44].
Considering that antibody-facilitated infection of phagocytic immune cells has been well characterised in SARS and MERS patients [45] and is associated with similar aberrant inflammatory responses in COVID-19 patients, antibody-facilitated enhancement of SARS-CoV-2 is quite possible. However, it is apparent that we still have more questions than answers for the new coronavirus. The role of the antibody response in SARS-CoV-2 infection is not well understood, and despite the temporal relationships between antibodies and cytokine-associated pathology, there are many other explanations that should not be excluded from consideration.
Selecting for Specificity
In all likelihood, infection with SARS-CoV-2 may elicit a combination of neutralising and enhancing antibodies – the amount, proportion and concentration of which shape a patient’s clinical outcomes. Therefore, attention should be paid to the nature of the immune response that a vaccine aims to induce.
The major determinant of whether an antibody is protective or enhancing is the epitope to which it is raised. Given the pivotal role of the Spike protein in mediating viral uptake and the immunodominant nAb responses it elicits [46], nearly all SARS-CoV-2 vaccines seek to target it [47]. However, while necessary to induce a protective response, Spike has also shown to be the key mediator of antibody enhancement [19]. As such, researchers are faced with a significant dilemma: formulation of Spike protein will be necessary to confer protective and sterilising immunity, but also carries the greatest risk of stimulating adverse events in patients and derailing vaccine development entirely.
To remedy this, researchers have explored various options. In vitro studies have shown that certain Spike epitopes tend to be more ‘dangerous’ than others in the types of antibodies they elicit [48]. Therefore, a popular vaccine design strategy has been to ablate cross-reactive and sub-neutralising epitopes, while attempting to retain the overall conformation of those that are protective. Generally-speaking, this is achieved by introducing random point mutations and screening for those which show the least affinity for sub-neutralising antibodies [49]. The most ‘neutralisable’ region of the Spike protein is its receptor-binding domain (RBD) within the S1 subunit, which incidentally happens to share the lowest sequence identity to other coronaviruses, making it a choice target for specific and effective vaccines [50].
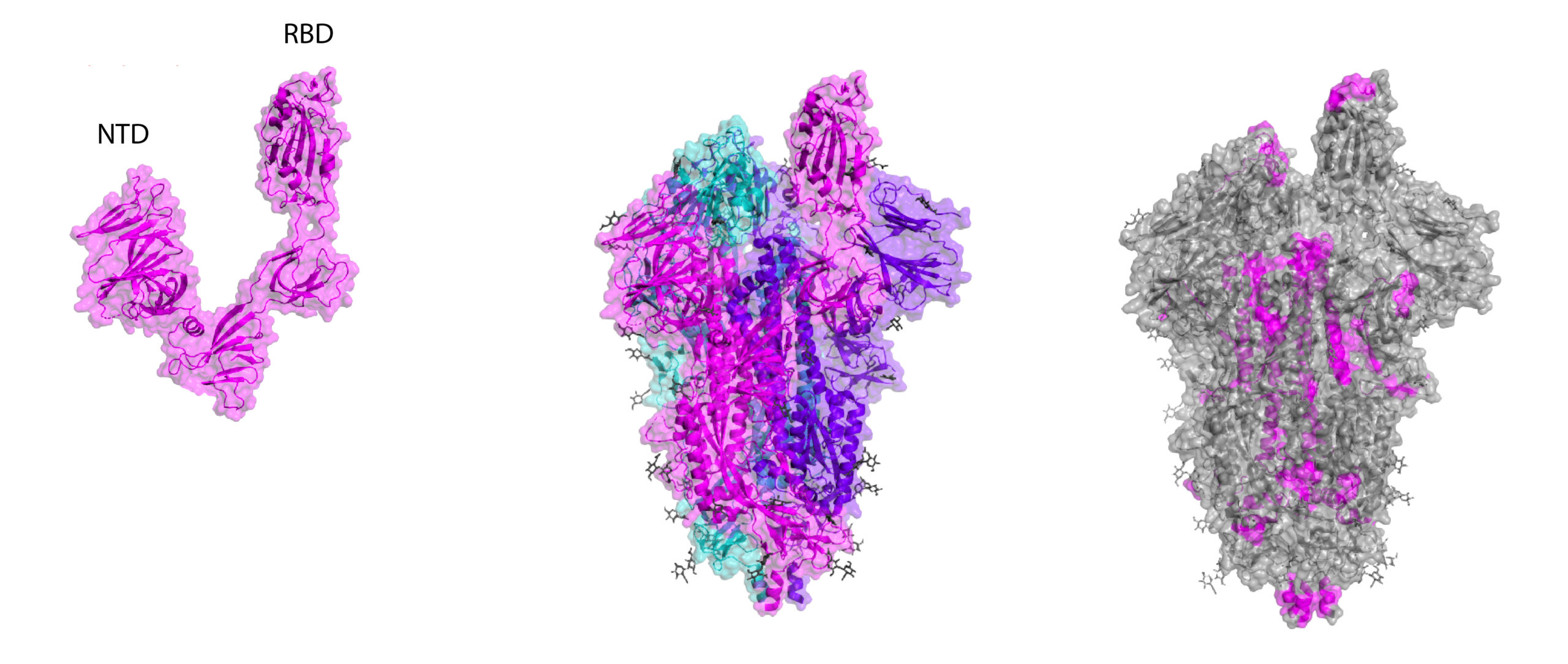
Crystal structures of RBD and whole Spike, including B-cell linear epitope map (right). Epitopes are taken from early homology modelling and bioinformatic studies by Ahmed and colleagues and Grifoni and colleagues.
However, vaccine design is never quite as straightforward as you might hope. A recent study of MERS Spike, for example, suggests that the RBD may be equally able to enhance disease [53]. Therefore, while a promising immunogen, more work is needed to mitigate the potential antibody-enhancing risks of Spike vaccines. Some of the areas that require further consideration are summarised below:
+ Non-linear epitope mapping studies, carried out in conjunction with the interrogation of enhancing peptides are needed to establish a comprehensive epitope map.
+ Spike trimers exists in multiple, distinct conformational states, before and after fusion with the host cell membrane [54, 55], and need to be assessed for their protective/enhancing effects accordingly.
+ While Spike subunits and peptides may mitigate the potential effects of ADE, it is not clear whether they would be sufficient in eliciting potent and protective nAb responses in vaccines.
+ Nucleoprotein is frequently formulated with Spike to broaden the immune response and is generally regarded as a ‘safer’ antigen. However, SARS Nucleoproteins have also shown to play a role in disease enhancement [56, 57], requiring thorough investigation of these formulations.
+ The seroprevalence of antibodies to the seasonal coronaviruses across different age groups and their ability to induce amnestic effects is not understood, and should be investigated in both serosurveys and vaccine trials [13].
+ The divergence in results from early in vitro and in vivo studies [58, 59] suggests that these models are not well suited to capture the nuances of the human immune system. Researchers should therefore tread with caution when making interpretations and extrapolations to humans.
+ Ancillary factors include the concentration, affinity and isotypes of antibodies elicited, and the effects of adjuvant and route of administration [60].
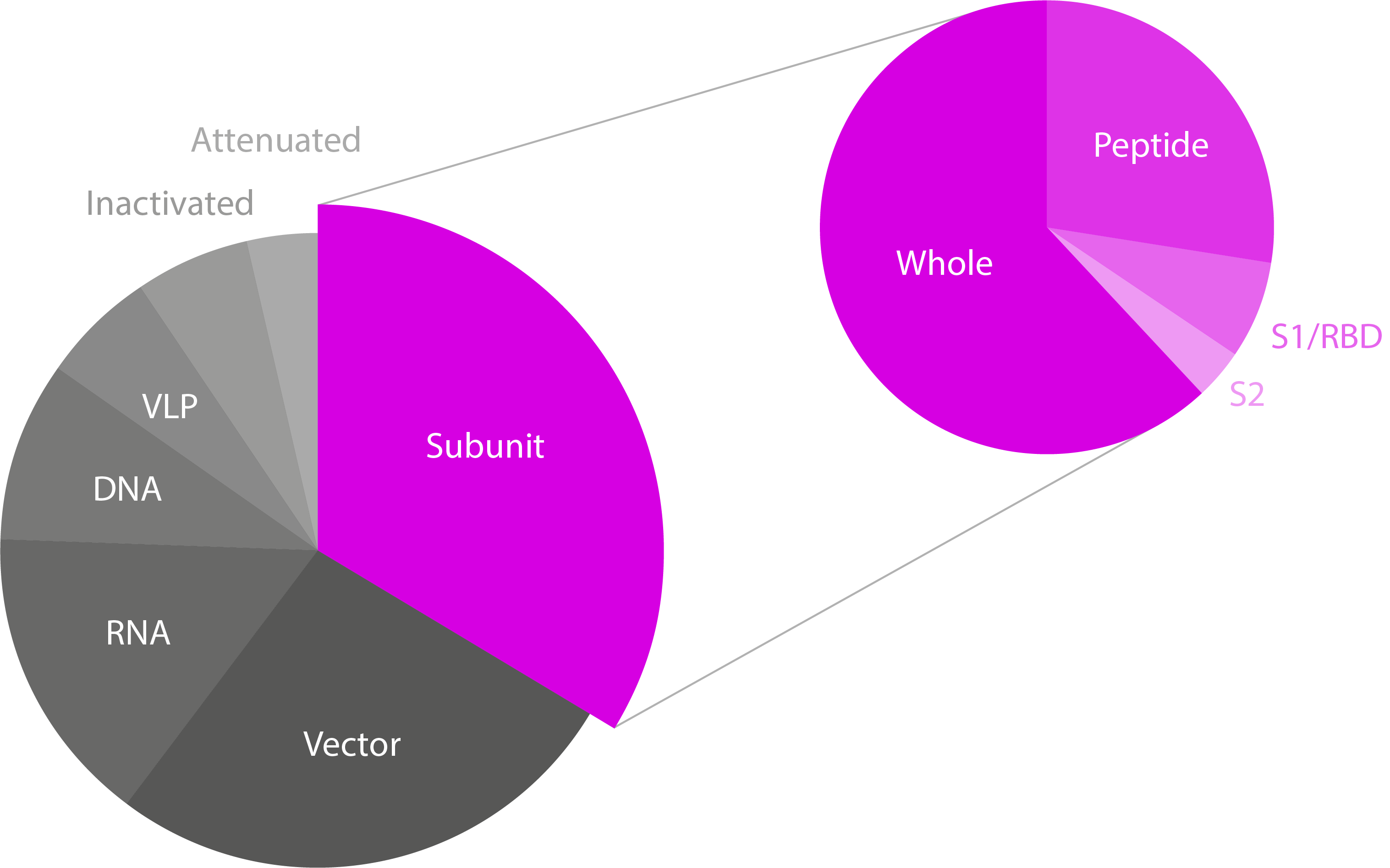
Pie charts showing the different SARS-CoV-2 vaccine technologies/platforms and the type of subunit vaccines in development. Note that only subunit vaccines have been highlighted, though the majority of candidates for all platforms are based on Spike. Data from WHO.
Reading the Response
Given that the quality and quantity of patient antibodies will largely dictate the functional outcomes of a vaccine, a final, yet nonetheless critical, consideration is the need to develop vaccine diagnostics. Specifically, vaccine candidates will require the development of bespoke immunoassays, each of which will require extensive calibration to reach the required specificity and sensitivity needed to provide robust datasets. The insights gained from such studies should, in turn, help to build a better picture of the correlates of protection vs enhancement and inform subsequent candidates.
When looking at the functional requirements, such assays should be able to measure indicators of ADE in vaccine recipients, including those who are subsequently infected, with pre- and post- measurements of anti-Spike/Nucleoprotein IgG, and the proportion of antibodies that are directed against ADE-associated epitopes [37]. In particular, multiplex assays that are able to characterise the ratio of useful nAbs to those that are enhancing, will prove instrumental in demonstrating safety. In addition, given the evidence of ADE at sub-neutralising antibody titres [61] and the likelihood of this in the context of vaccination, assays will also need to be highly sensitive for detection of nAbs at low levels.
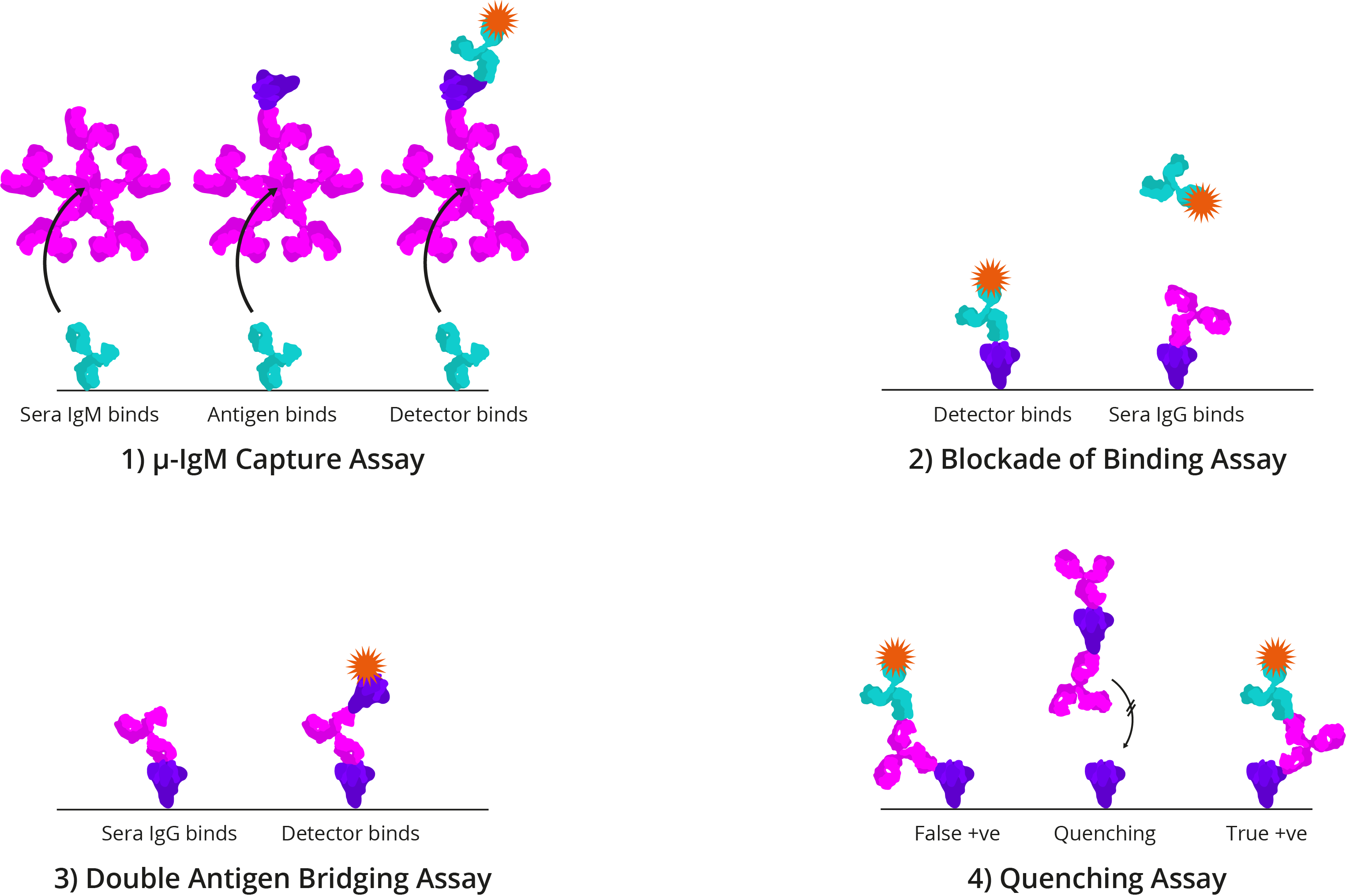
Four alternative ELISA formats: 1) μ-IgM capture assay: IgM antibodies in patient sera bind to well-bound anti-human IgM. Once bound, Spike and detection antibody bind; 2) Blockade of binding assay: In the absence of patient antibody sera, a specific monoclonal antibody binds well-bound Spike and produces a signal. In the presence of patient antibody sera, patient antibodies bind well-bound Spike and exclude the detection antibody, resulting in a negative signal; The monoclonal antibody utilised binds a highly specific Spike epitope, and thus is only blocked by CoV-specific patient sera and not cross-reactive CoV sera; 3) Double antigen bridging assay: Patient sera antibody binds well-bound Spike, followed by labelled detection Spike; This format inherently detects higher affinity antibodies with higher specificity; 4) Quenching assay: Non-specific CoV patient sera antibodies are able to bind cross-reactive Spike epitopes leading to false positive results. To prevent this, excess Spike is added in solution to quench cross-reacting antibodies. This allows only CoV-specific antibodies to produce a true-positive signal.
Under normal circumstances, vaccine candidates would take years to mature from concept to entering clinical trials. Yet the urgency with which a vaccine for COVID-19 is needed will likely compress these timelines significantly. However, in our haste to find a solution, we mustn’t rush forward blindly. As a recent dengue vaccine has shown, antibody-enhancing effects can not only increase mortality in those susceptible, but open severe disease to new age groups and undo years of progress in building public confidence. Similarly, the coronaviruses don’t have a great track record when it comes to vaccines, and it would not be wrong to assume that the trials and tribulations for SARS-CoV-2 could be similar. Therefore, combined with the fact that COVID-19 is not life-threatening to much of the global population, toleration of side-effects by regulators will be low.
The Right Reagents
Crucial to the development of an effective diagnostic assay are the choice of quality critical reagents used to design it. Incidentally, Spike is a highly complex protein, whose trimers contain up to 66 N-linked glycans that facilitate protein folding, improve stability, alter viral tropisms, evade viral recognition, and block access of proteolytic enzymes during infection [62]. From the perspective of assay design, these glycans constitute many of the key surface epitopes of Spike that are recognised by antibodies in assays. Conversely, the use of Spike proteins with unglycosylated sequons risk the binding of non-specific, cross-reactive antibodies that risk inducing false-positive results. Therefore, assay developers should take great care in their choice of reagents.
At The Native Antigen Company, our proprietary VirtuE (HEK293) expression system ensures complete glycosylation and proper folding of our coronavirus antigens, to ensure full biological and antigenic activity for the development of immunoassays. For more information about our reagents, please see our antigen and antibody ranges below:
You can keep up-to-date with new product developments on our Coronavirus Pipeline page. If we are not currently developing something that you’re interested in, please get in touch at contact@thenativeantigencompany.com, as we may be able to develop it for you.
References
1. https://www.worldcat.org/title/roitts-essential-immunology/oclc/949912303
2. https://www.liebertpub.com/doi/pdf/10.1089/088282403763635465
3. https://pubmed.ncbi.nlm.nih.gov/26497532/
4. http://dx.doi.org/10.1128/JVI.64.3.1407-1409.1990
5. https://link.springer.com/article/10.1007%2FBF01310476
6. https://www.ncbi.nlm.nih.gov/pubmed/17382365
9. https://thenativeantigencompany.com/an-early-look-at-vaccines-for-covid-19/
10. https://insight.jci.org/articles/view/123158
11. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0035421
12. https://www.ncbi.nlm.nih.gov/pubmed/12781535
13. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3367364/
14. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016775/
15. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7112410/
16. https://www.ncbi.nlm.nih.gov/pubmed/24885320?dopt=Abstract
17. https://www.ncbi.nlm.nih.gov/pubmed/21775467?dopt=Abstract
18. https://pubmed.ncbi.nlm.nih.gov/15602737/
19. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6478436/
20. https://www.ncbi.nlm.nih.gov/books/NBK349040/
21. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5574614/
22. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7022351/
23. https://coronavirus.jhu.edu/map.html
24. https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30243-7/fulltext
25. https://www.jto.org/article/S1556-0864(20)30132-5/pdf
26. https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciaa248/5803306
27. https://www.ncbi.nlm.nih.gov/pubmed/12734147
28. https://www.ncbi.nlm.nih.gov/pubmed/12682352
29. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3367364/
30. https://www.thelancet.com/journals/lanres/article/PIIS2213-2600(20)30216-2/fulltext
31. http://apjai-journal.org/wp-content/uploads/2020/02/Covid_AP-200220-0772.pdf
32. https://www.nejm.org/doi/full/10.1056/NEJMsr1408795
33. https://www.medrxiv.org/content/10.1101/2020.03.30.20047365v1
34. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3367364
35. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016775/
36. https://www.medrxiv.org/content/medrxiv/early/2020/03/04/2020.03.03.20028423.full.pdf
38. https://www.nature.com/articles/s41418-020-0530-3
39. https://www.medrxiv.org/content/10.1101/2020.02.19.20024885v1.full.pdf
40. https://www.medrxiv.org/content/10.1101/2020.03.27.20045427v1
41. https://www.biorxiv.org/content/10.1101/2020.02.12.945576v1
42. https://www.medrxiv.org/content/10.1101/2020.02.16.20023671v1.full.pdf+html
43. https://www.medrxiv.org/content/10.1101/2020.02.18.20024364v1.full.pdf
44. https://www.frontiersin.org/articles/10.3389/fimmu.2020.00827/full
45. https://www.sciencedirect.com/science/article/pii/S004268221400049X?via%3Dihub
46. https://www.biorxiv.org/content/10.1101/2020.04.23.056853v1
47. https://www.who.int/who-documents-detail/draft-landscape-of-covid-19-candidate-vaccines
48. https://pubs.acs.org/doi/10.1021/acsinfecdis.6b00006
49. https://www.microbiologyresearch.org/content/journal/jgv/10.1099/vir.0.82640-0?crawler=true
50. https://link.springer.com/article/10.1007%2Fs11427-020-1637-5
51. https://www.mdpi.com/1999-4915/12/3/254
52. https://www.sciencedirect.com/science/article/pii/S1931312820301669
53. https://jvi.asm.org/content/94/5/e02015-19
54. https://journals.plos.org/plospathogens/article?rev=2&id=10.1371/journal.ppat.1007236
55. https://www.sciencedirect.com/science/article/pii/S0092867418316428
56. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4561025/
57. https://www.jimmunol.org/content/181/9/6337
58. https://www.ncbi.nlm.nih.gov/pubmed/21775467?dopt=Abstract
59. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7115629/
60. https://www.nature.com/articles/nrmicro2090
61. https://www.ncbi.nlm.nih.gov/pubmed/25073113
62. https://www.biorxiv.org/content/10.1101/2020.03.26.010322v1.full.pdf

