During the course of the current coronavirus pandemic, we have all been aware of the urgent need for nucleic acid testing to identify people currently infected with SARS-CoV-2. The second form of testing needed are serological immunoassays, which can identify past infections but are more complex to develop. In this Q&A, Dr Andy Lane discusses adaptive immunity and the production of antigens and antibodies for the development of immunoassays for COVID-19 testing. This article was originally published by Clinical Laboratory Inc.
Can you tell me about the company and its history?
The Native Antigen Company was founded in Oxford, UK in 2010, with the goal of developing native viral and bacterial antigens to support the in vitro diagnostics industry. Following its success in developing reagents during the 2015/2016 Zika epidemic, the company underwent a period of rapid growth and began producing broader range of native and recombinant antigens, as well as offering custom and contract services to the life sciences and biotechnology industries. Fast forward to February of this year, and the Company became one of the first-recognised suppliers of antigens for SARS-CoV-2, and has since developed a broad and expanding range of coronavirus reagents to support ongoing research and development.
Our reagents are used by a wide-range of researchers working in infectious diseases, but are predominantly sold into two major markets: The in vitro diagnostics (IVD) industry, who use antigens and antibodies to develop immunoassays for serological diagnosis of infection, and the vaccine industry, who use antigens and antibodies to develop immunoassays for the qualification and quantification of animal and patient vaccine responses in clinical trials.
Can you provide an overview of how immunity is generated in response to infection?
It goes without saying that the human immune system is highly complex, but it can generally be broken down into the innate and adaptive immune responses: Innate immunity is our first line of defence. It provides a rapid, but somewhat makeshift response that is largely preoccupied with trying to kill infectious agents from the moment they enter the body with a broad array of non-specific cells, proteins and biochemicals. While this is ongoing, the innate response alerts the adaptive response. Adaptive immunity represents the elite troops of the immune system, which launch an attack that is specifically adapted to the infectious agent using more sophisticated weapons to mediate powerful downstream responses. The hallmark of the adaptive response is clonal expansion, where B and T lymphocytes that are able to recognise a pathogen will be positively selected for to rapidly build their numbers. Once these cells reach significant levels, the body is much better equipped to detect and clear the invading pathogen, and tends to form a long-lasting ‘memory’ of the pathogen to better prepare itself for future encounters.
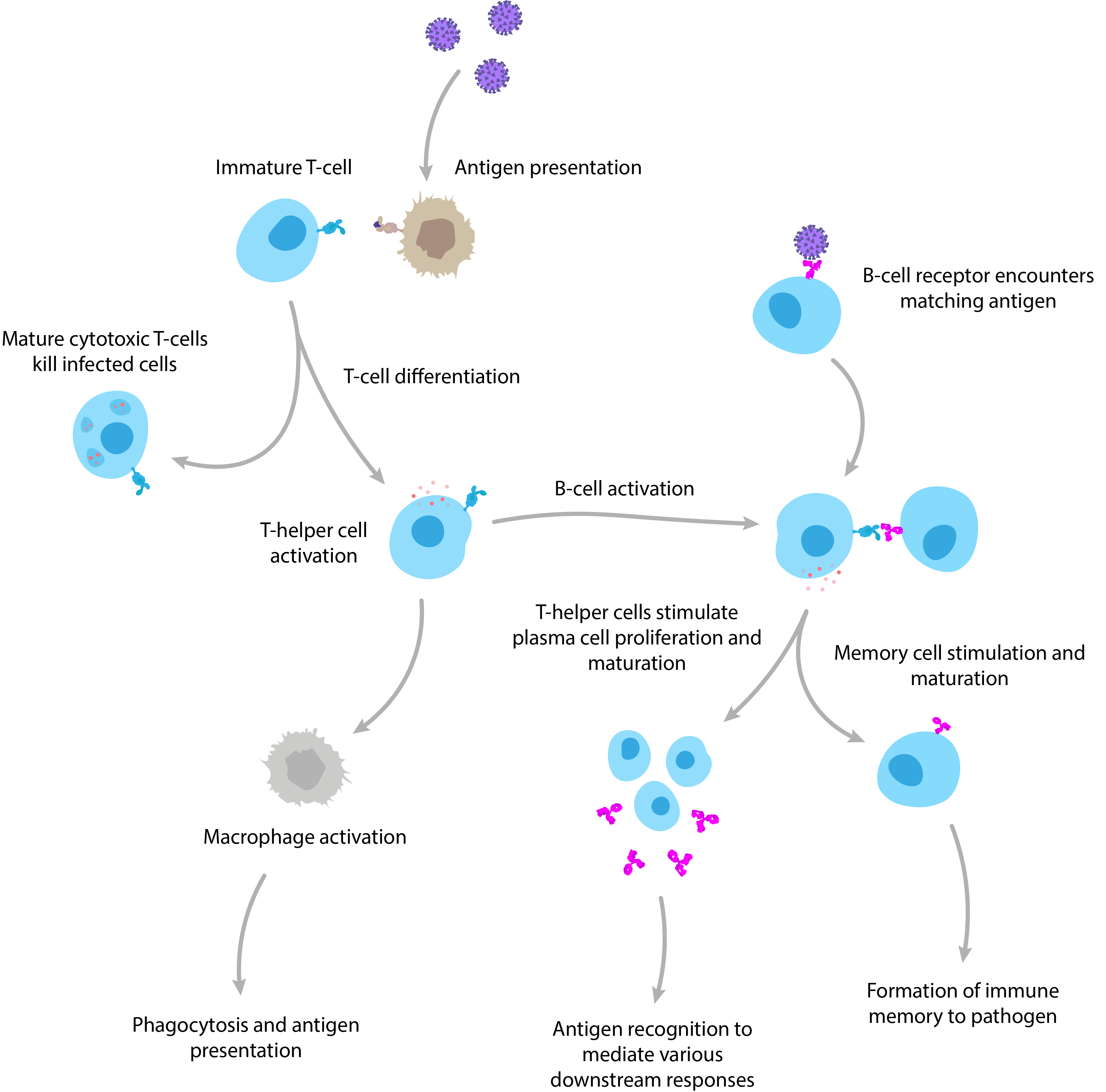
A simplified diagram of adaptive immunity showing T-cell and B-cell activation and function.
After some viral infections, we never get them again, but after others we are only protected for a short period of time – why does this difference arise?
There are two major reasons for reinfection by a virus, shortly after initial exposure. The first is due to the ability of viruses to mutate, which occurs via the natural accumulation of genetic changes over time (antigenic drift) or recombination of a virus’s genome with a related strain, causing it to rapidly mutate into a novel form (antigenic shift). These processes allow the virus to change its ‘appearance’, such that it is no longer recognisable by our immune system, and makes our previous exposure to the original virus of little use. This is best exemplified by the influenza A virus, which is notorious for mutating its surface proteins (hemagglutinins and neuraminidases) to evade immune recognition, resulting in a perpetual game of cat and mouse that requires the development of new vaccine formulations every flu season.
The second reason for ineffective immune responses is a bit more complex and tends to occur as a result of waning memory cell levels in the host’s immune system following the initial infection. However, the cause of short-lived immunity is not entirely clear and largely depends on the virus in question as well as myriad influencing factors, such as genetics, age and previous exposure to pathogens. A very relevant example are the endemic coronaviruses, such as OC43-CoV and 229E-CoV, whose infections may result in only a few months of immunity. A study in the early 90s, for example, showed that infection with 229E-CoV only one year after initial exposure resulted in reinfection in the majority of patients and correlated with declining antibody titres [1]. The reason for the decline in immune memory is not entirely clear but is often attributed to the mild pathogenicity of such viruses eliciting a somewhat lacklustre immune response in the first place.
Given the short-lived immunity of some coronaviruses, COVID-19 immunity has been a hot topic. Most patients have shown quite potent and lasting antibody responses, while some have little-to-no detectable antibodies following infection [2]. While we are not yet sure whether this is an immune phenomenon or an issue with assay insensitivity, it will take some time before we are able to truly understand our body’s response to this disease.
Serology testing is of great importance in clinical diagnostics. When doing serology testing to see if a person has had a disease, what exactly is being detected and how is this usually achieved?
By definition, serology is the scientific examination of blood serum and its components. However, in the context of the clinical diagnosis of infectious disease, it generally refers to the use of immunoassays that measure antigens or antibodies. Immunoassays are found in a wide variety of formats, but are best exemplified by the enzyme-linked immunosorbent assays (ELISA), which use plastic titre plates to bind antigens or antibodies from patient samples and produce a detectable signal.
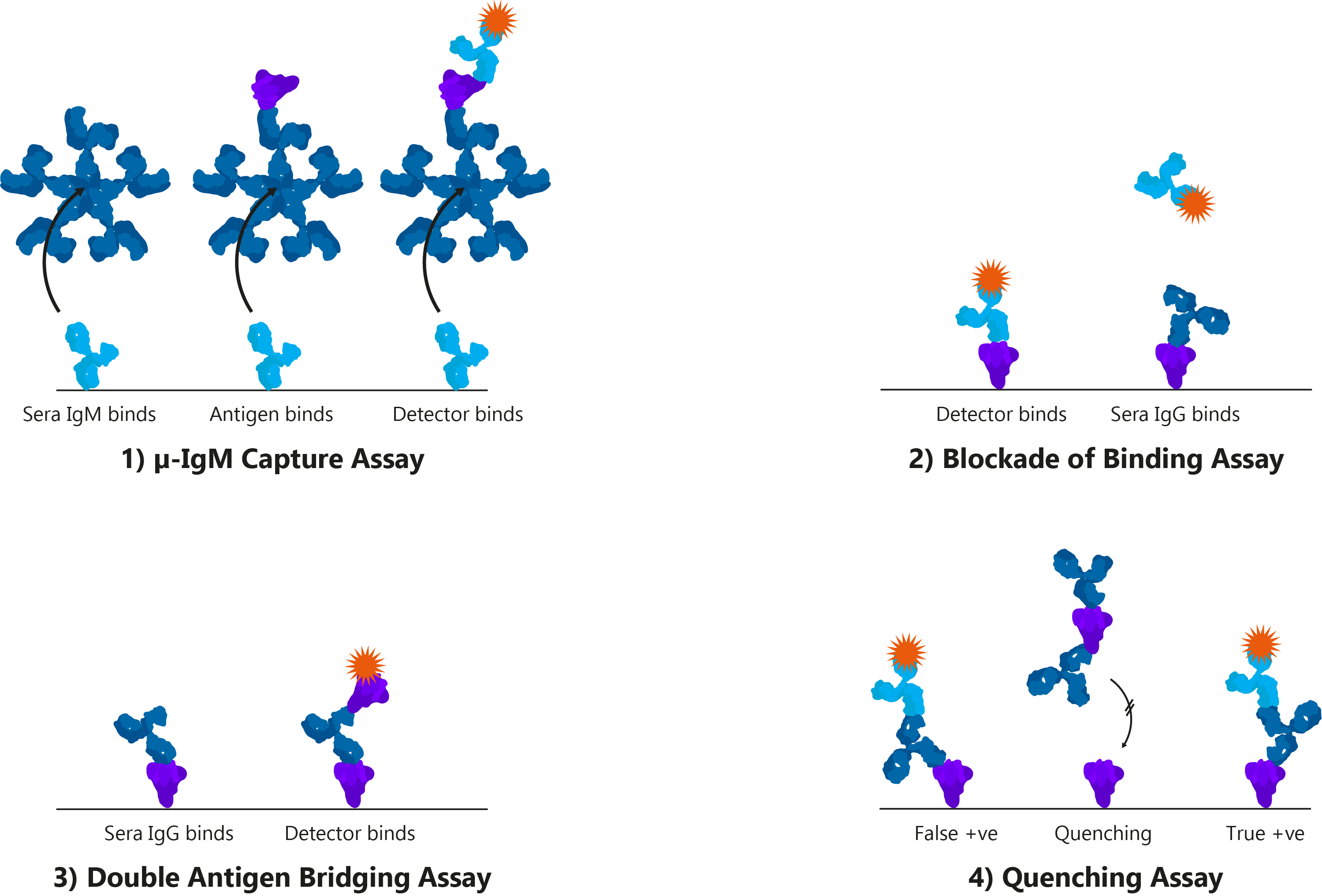
Four alternative ELISA formats: 1) μ-IgM capture assay: IgM antibodies in patient sera bind to well-bound anti-human IgM. Once bound, Spike and detection antibody bind; 2) Blockade of binding assay: In the absence of patient antibody sera, a specific monoclonal antibody binds well-bound Spike and produces a signal. In the presence of patient antibody sera, patient antibodies bind well-bound Spike and exclude the detection antibody, resulting in a negative signal; The monoclonal antibody utilised binds a highly specific Spike epitope, and thus is only blocked by CoV-specific patient sera and not cross-reactive CoV sera; 3) Double antigen bridging assay: Patient sera antibody binds well-bound Spike, followed by labelled detection Spike; This format inherently detects higher affinity antibodies with higher specificity; 4) Quenching assay: Non-specific CoV patient sera antibodies are able to bind cross-reactive Spike epitopes leading to false positive results. To prevent this, excess Spike is added in solution to quench cross-reacting antibodies. This allows only CoV-specific antibodies to produce a true-positive signal.
The second major immunoassay format is the lateral flow assay (LFA), which use an absorbent pad to absorb and analyte and run it though a series of specific antibodies to produce a detectable signal. These assays have the advantage of being inexpensive and portable and can typically provide results within minutes.
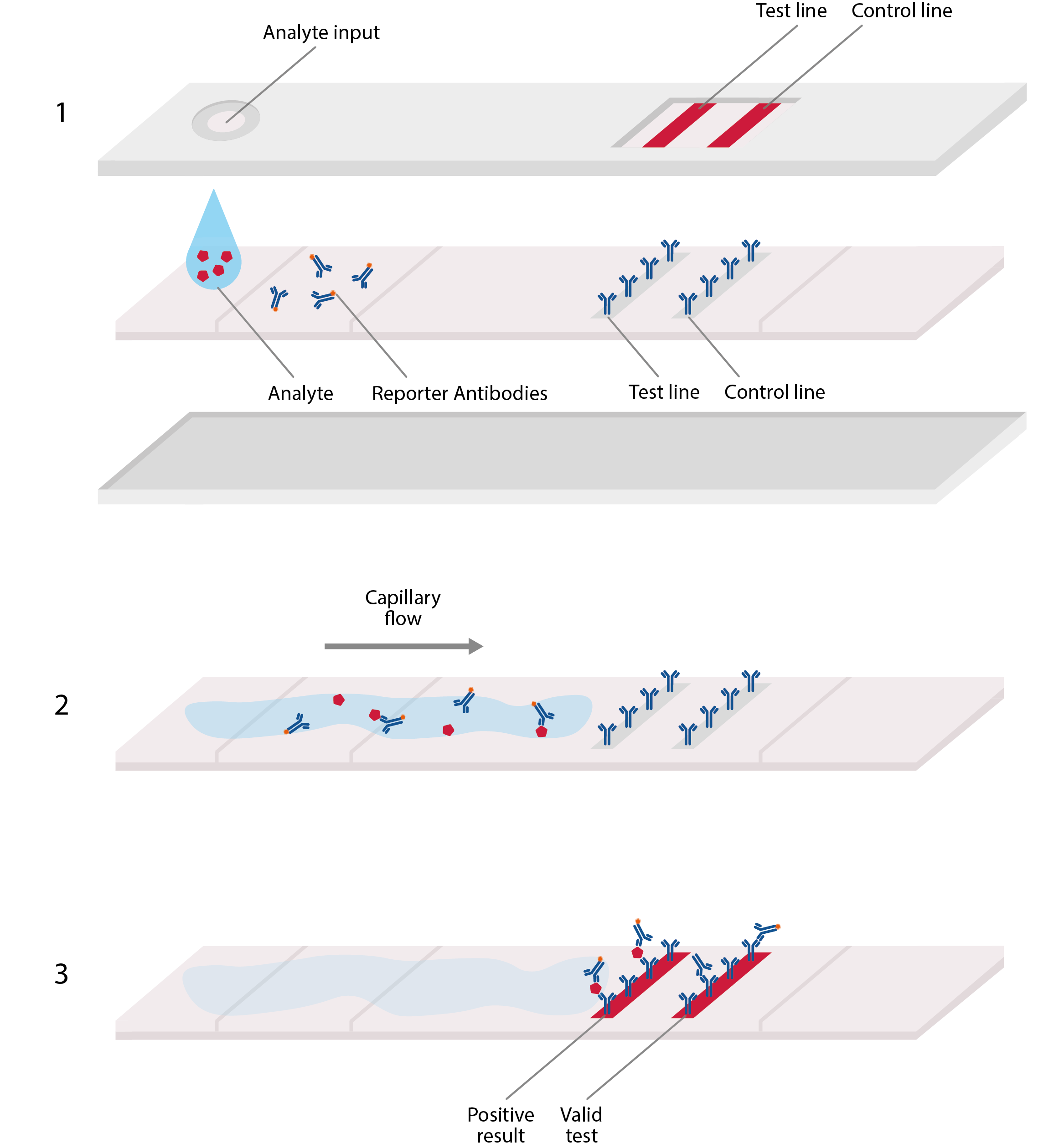
Basic design of lateral flow assay, akin to home pregnancy tests. LFAs typically comprise a plastic cassette, containing a strip of paper that is able to absorb and transport analyte. 1) Analyte from blood or sputum is first added to one end of the paper strip, where it is absorbed by the paper, allowing the viral antigens to migrate along it. 2) These antigens first encounter an area of conjugate antibodies, which adhere to them, along with detectable tags. 3) The antigens then migrate to an area of test antibodies that are highly specific to the virus and bind to produce a solid, visible line that indicates a positive result, and hence, confirmed infection. Secondary control lines can then be used to detect conjugate antibodies and ensure that the test is working correctly.
Emerging studies suggest that the serology of SARS-CoV-2 is highly complex and differs significantly from other betacoronaviruses. Antibody responses to SARS-CoV-2 appear to occur later and be of lower titres than are typically observed for viral infections, influencing the way in which assays are designed to diagnose both acute and historic infections. Another important consideration is the potential for antibody cross-reactivity to other co-circulating coronaviruses, requiring close attention to the binding specificity of antigens used.
In the current COVID-19 pandemic, serology testing will be crucial for discovering much about the disease – what will we be hoping to learn from this?
From the outset of the pandemic, the reverse-transcriptase polymerase chain reaction (RT-PCR) has been the predominant means of diagnosing active infection. However, as molecular methods rely on the presence of viral nucleic acids, they are limited to a narrow window during the acute phase of infection when virus is present in the respiratory tract. This has left a major gap in the ability to detect previous cases and understanding the transmission dynamics of this disease. On the other hand, antibodies to SARS-CoV-2 may last for some time after infection to allow for retrospective diagnosis once patients have recovered. This is particularly useful for multiple reasons.
First, as governments ease lockdowns, high quality epidemiological data is vital to keeping an eye on temporal and geographical disease dynamics, which will require frequent sampling of antibodies in populations (serosurveys). There is also a clear advantage in using serology tests for diagnosis at the point-of-care (PoC). Unlike high-throughput RT-PCR or ELISA immunoassays, LFAs present a highly practical and rapid alternative for acute-phase diagnosis and will be crucial in identifying asymptomatic carriers and infected individuals to ensure they are isolated from the general population.
Another major role of serology is in vaccine testing. So far, there are over 130 vaccine candidates currently in the pipeline [3]. While these vaccines are based on a wide range of platforms, (including mRNA, DNA, nanoparticles, subunits, synthetic peptides and virus-like particles, to name a few), it can be said with near certainty, that a SARS-CoV-2 vaccine will elicit immune responses to the Spike protein. However, considering that vaccine-induced anti-Spike IgG levels may be indistinguishable from those conferred by natural infection, alternative antigens will be needed to design vaccine-specific assays. These assays will also be very useful in assessing the potential risk of vaccine-induced antibody-dependent enhancement (ADE), in which antibodies produced by a vaccine are able to facilitate a more aggressive pathogenesis when a patient gets a real SARS-CoV-2 infection.
How do you go about preparing reagents for a serology test for a new pathogen such as SARS-CoV-2 and why is it important that these reagents are ‘native-like’?
When developing any immunoassay, the most important components are the antigens and antibodies used to design it. The considerations for choosing these reagents are wide-ranging: antigens should include the most appropriate epitopes to facilitate high sensitivity and antibodies should be tested for high affinity to the antigen in question. When considering specificity, it is crucial to ensure than detector antibodies do not bind to the cross-reactive epitopes that are often found on more conserved regions of viral antigens.
To modulate the sensitivity and specificity of an assay, specific portions of a protein can also be used. In the case of SARS-CoV-2, researchers are investigating various different regions of its Spike protein for use in immunoassays. The S1 and S2 subunits of Spike are a popular choice for the development of immunoassays as they are highly exposed to the virus’s external environment and can readily induce potent antibody responses. In particular, Spike antibodies that bind the RBD of S1 may be able to neutralise virus by preventing binding with ACE2. The Spike receptor-binding domain (RBD) functions to mediate cell-surface attachment and internalisation by binding human ACE2 receptors. Given RBD’s role in host-cell entry, it is able to elicit highly neutralising antibody responses and is a popular target for the development of vaccines. The RBD also shows high sequence divergence between other coronavirus Spike proteins, making it a popular antigen for the development of sensitive and specific immunoassays. The N-terminal domain of the SARS-CoV-2 Spike protein shows the highest sequence variability across the coronavirus family, making it a popular choice of antigen for maximising the specificity of diagnostic assays.
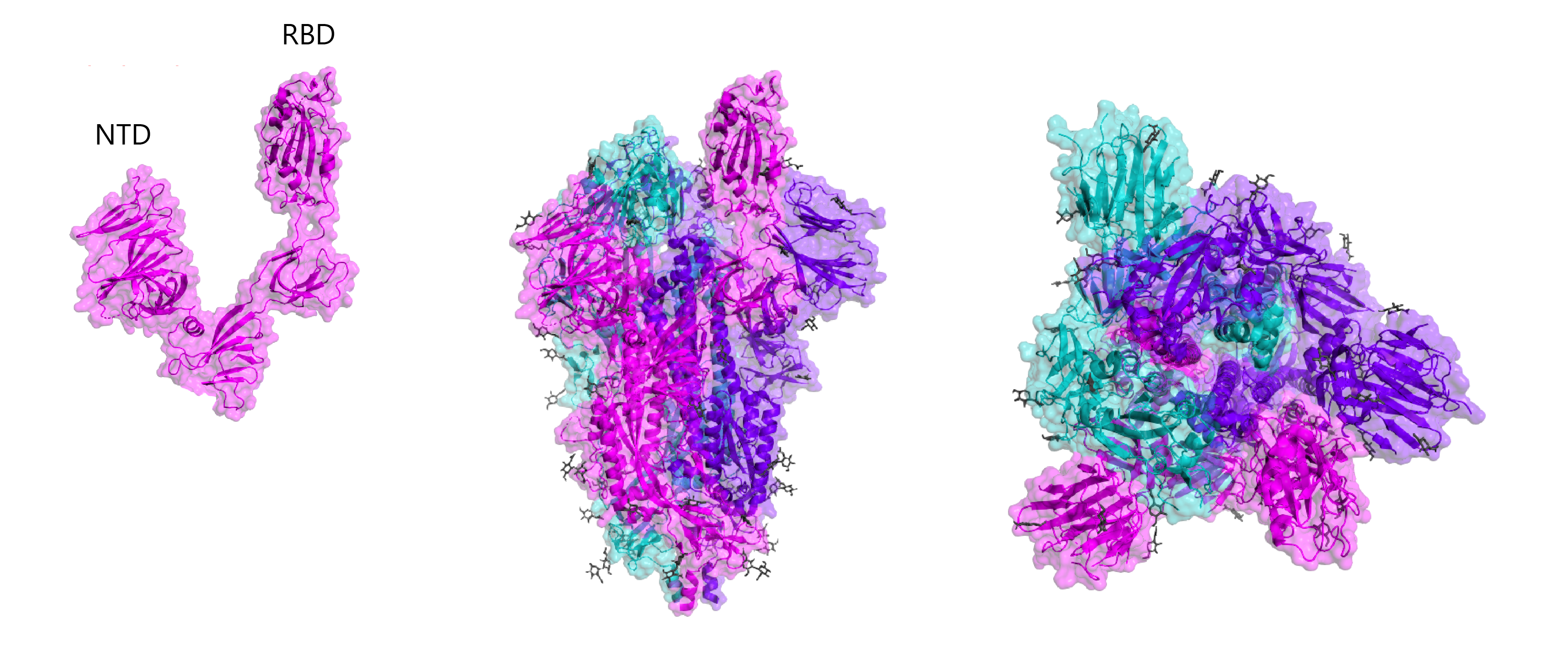
Crystal structures of RBD and whole Spike, including B-cell linear epitope map (right). Epitopes are taken from early homology modelling and bioinformatic studies by Ahmed and colleagues and Grifoni and colleagues.
Given the biosafety implications of handling live virus, recombinant antigens expressed from other organisms are the go-to for developing assays. However, not all expression systems are born equal. Simple organisms like E. coli are easy to genetically manipulate but lack the necessary post-translational machinery to glycosylate proteins. Incidentally, each SARS-CoV-2 Spike trimer contains up to 66 glycan sugars (see figure above) to facilitate folding and mediate viral tropisms, amongst other things. From the perspective of assay development, these glycans constitute many of the key surface epitopes that are recognised by detector antibodies and the use of unglycosylated Spike risks the binding of non-specific, cross-reacting antibodies that can reduce diagnostic specificity.
To ensure that Spike is produced with its full glycosylation pattern and is properly folded, more complex systems need to be used. At The Native Antigen Company, we use our VirtuE mammalian (HEK293) system that has been developed for the bespoke purpose of expressing high quality antigens with proper folding and full glycosylation.
What’s your vision for the future for The Native Antigen Company and its collaboration with OXGENE?
After the SARS-CoV-2 genome was published in early January, it was an all-out race to develop and release reagents. After a tremendous effort by our R&D team, we managed to produce our first batch of S1 antigens in early February and began to ship them to our customers around the globe. However, the next challenge was manufacturing capacity. Given the demand from the IVD and vaccine industries, we soon began to struggle in meeting such large demand. Fortunately, we were able to reach out to some manufacturers who could support us with scale production.
Our first partner, OXGENE™ have been using their Protein Machine Technology to develop stable cell lines for the production of Spike antigens. Their technology uses a proprietary adenoviral vector to carry SARS-CoV-2 DNA into human cells, where it delivers it to the nucleus for stable integration. From here, cell lines can be cultured en masse to produce large quantities of protein without the inherent limitations in yield of transient expression. Work is still ongoing to optimise expression, but we’re hoping for some positive data in the coming weeks.
References
1) Callow, KA., Parry, HF., Sergeant, M., Tyrrell, DA. (1990). The time course of the immune response to experimental coronavirus infection of man. Epidemio Infectl 105(2); 425-446
2) Immune responses and immunity to SARS-CoV-2 [internet]. European Centre for Disease Prevention and Control 2020, 19 June (https://www.ecdc.europa.eu/en/covid-19/latest-evidence/immune-responses).
3) Draft landscape of COVID-19 candidate vaccines [internet]. World Health Organization 2020, 18 June (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines).