Enabling a new generation of Human Immunodeficiency Virus 1 (HIV-1) and HIV-2 diagnostic assays and structural studies
Sensitive and accessible diagnostics and medication remain the only tools to tame the spread and protect the population from the life-limiting acquired immunodeficiency syndrome (AIDS). Human immunodeficiency virus 1 (HIV-1) is the most common cause of AIDS worldwide, while HIV-2 infection is confined mainly to the population of Western Africa. The clinical characteristics of those HIV infections differ as the patients infected with HIV-1 are more likely to progress to AIDS without sufficient treatment and early diagnosis (Nyamweya et al., 2013).
The viral entry receptor for HIV-1 and HIV-2 is the Envelope protein gp160, a transmembrane glycoprotein featuring the mature glycoprotein gp120 and the transmembrane protein gp41. Both neutralising and non-neutralising patient antibodies are targeted against gp160, making this protein an exciting diagnostic and vaccine target. gp160 is synthesised as a polyprotein and cleaved by the host-resident protease furin (reviewed in Checkley et al., 2011).
Removal of the intravirion and transmembrane domains allow for production of a soluble secreted version of gp160 termed gp140. The Native Antigen Company (TNAC) now offers untagged HIV-1 glycoprotein gp140 group M subtypes A, B, C, CRF01-AE, group O, and HIV-2 glycoprotein gp140, enabling a new generation of diagnostic assays and structural studies. All proteins are modified using three amino acid changes (generally referred to as SOSIP) so that a disulfide bridge covalently links gp120 to the soluble domain of gp41, termed gp21, to increase the stability and longevity of the complex (Binley et al., 2000).
Classification and Epidemiology
The transmissibility of HIV-1 can be attributed to the extensive genetic heterogeneity of the virus, resulting in the generation of many variants of this virus with varying global distribution. Four major phylogenetic groups of the HIV-1 variants include the Main group (M), Outlier group (O), non-M or O group (N), and most recently added group P (Vallari et al., 2011).
Most HIV-1 infections worldwide are associated with the Main group, divided into A-K subtypes. The groups O, N, and P are estimated to be responsible for only about 1% of all HIV-1 cases, while the subtypes of group M are associated with approximately 95% of the total HIV-1 burden (Giovanetti et al., 2020).
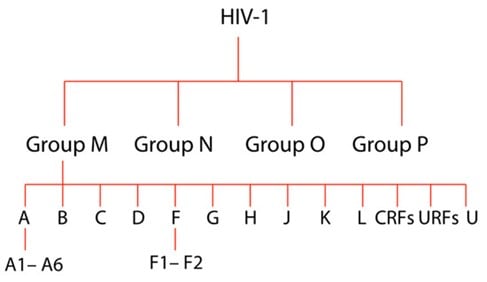
Figure 1. Graphical representation of HIV-1 evolutionary relationships between HIV-1 strains. From the top, there are four main groups, subtypes (A-U) and sub-subtypes (for example, A1-A-6), where ‘U’ signifies the untyped yet ‘CRFs’ is an abbreviation of circulating Recombinant Forms and ‘URFs’ means Unique Recombinant Forms.
Subtype C is the most prevalent form of the virus, primarily associated with disease cases from low-income countries like India or Ethiopia and Southern Africa, where 1 in 4 adults live with HIV-1 infection. Although disproportionately higher, publicity and research interest is associated with the HIV-1 subtype B, which is most prevalent in Europe and the Americas, it accounts for only about 12% of all HIV-1 infections (Hemelaar et al., 2019).
Subtype CRF01_AE and several other CRFs are the main HIV-1 subtypes in East and Southeast Asia. The rapid emergence of various CRFs in this region began during co-circulation of the subtypes B and CRF01_AE in different risk groups. As a result, other more complex recombinant strains emerged from these subtypes, such as CRF15_01B11 and CRF34_01B, initially found in Thailand, Malaysia, and Singapore (Liu et al., 2012).
Viral Invasion
HIV-1
The viral envelope protein gp160 is central for HIV-1 invasion, enabling membrane fusion and subsequent genetic core delivery to the host cell.
When the HIV Envelope protein makes contact with the host cell membrane, it binds to the host receptor CD4 and coreceptors CCR5/CXCR4. This binding triggers a multistep conformational change in the envelope glycoprotein trimer. Those conformational changes lead to fusion peptide exposure allowing subsequent viral-host cell membrane fusion and entry of the genome into the host cell cytoplasm (Wang et al., 2020).
Natural noncovalent interactions between the six subunits of the HIV-1 envelope glycoprotein complex cannot maintain the trimeric structure of recombinantly expressed proteins. TNAC customers reported that the recombinant gp41 antigen alone is typically insoluble and difficult to handle without buffers containing high concentrations of ionic detergents, making it poorly suited to IVD assay manufacturing.
Our Solution to the HIV Assay Manufacture Challenges
A disulfide bond is introduced between gp120 and gp41 to stabilise the native-like structure of the HIV-1 glycoprotein complex, providing a bridge between gp120 and the gp41 soluble domain, gp21. The resulting gp140 antigens (SOSIP) mimic the antigenic properties of the native HIV envelope proteins, which, as the main target of the neutralising antibody responses, are critical in assay development and vaccine design studies (Pugach et al., 2015). The recombinant gp140 protein is soluble and stable in simple physiological buffers for convenient assay development and production (Harris et al., 2011).
Through altering the amino acid sequence of the native furin cleavage site, we promoted enhanced cleavage of the gp140 polyprotein, resulting in optimal presentation of the gp120:gp21 ectodomain antigenic target as described in Binley et al., 2000.
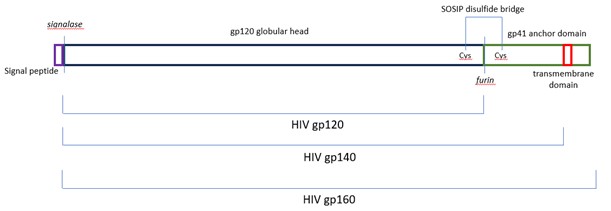
Figure 4. Our gp140 (SOSIP) feature an enhanced furin cleavage site that allows for maturation of the proteins (Binley et al., 2000) and a disulfide bond between gp120 and gp21, to remain covalently linked (Pugach et al., 2015).
The Native Antigen Company (TNAC) is pleased to announce launch of HIV-1 and HIV-2 gp140 (SOSIP) antigens of the following viral groups and subtypes:
- Human immunodeficiency virus subtype A, gp140 (SOSIP), untagged
- Human immunodeficiency virus subtype C, gp140 (SOSIP), untagged
- Human immunodeficiency virus subtype AE, gp140 (SOSIP), untagged
- Human immunodeficiency virus subtype B, gp140 (SOSIP), untagged
- Human immunodeficiency virus type 2, gp140 (SOSIP), untagged
- Human Immunodeficiency virus clade O, gp140 (SOSIP), untagged
To learn more about our comprehensive HIV antigen range please visit here:
At The Native Antigen Company, we also offer a variety of HIV antibodies to further support your R&D:
Can’t find what you are looking for?
The Native Antigen Company has experience working with world leaders in infectious diseases to meet their custom research and development needs. We are renowned for our agility and rapid turnaround time, delivering from concept to full-scale production within 8 weeks*. Click here to learn more about our custom services:
*Based on average turnaround times. More complex custom services may take longer.
References
- Binley, J.M. et al. (2000) ‘A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and GP41 subunits is an antigenic mimic of the trimeric virion-associated structure’, Journal of Virology, 74(2), pp. 627–643. doi:10.1128/jvi.74.2.627-643.2000.
- Checkley, M.A., Luttge, B.G. and Freed, E.O. (2011) ‘HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation’, Journal of Molecular Biology, 410(4), pp. 582–608. doi:10.1016/j.jmb.2011.04.042.
- Gartner, M.J. et al. (2020) ‘Understanding the mechanisms driving the spread of subtype C HIV-1’, EBioMedicine, 53, p. 102682. doi:10.1016/j.ebiom.2020.102682.
- Giovanetti, M. et al. (2020) ‘Molecular epidemiology of HIV-1 in African countries: A comprehensive overview’, Pathogens, 9(12), p. 1072. doi:10.3390/pathogens9121072.
- Harris, A. et al. (2011) ‘Trimeric HIV-1 glycoprotein GP140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures’, Proceedings of the National Academy of Sciences, 108(28), pp. 11440–11445. doi:10.1073/pnas.1101414108.
- Hemelaar, J. et al. (2019) ‘Global and regional molecular epidemiology of HIV-1, 1990–2015: A systematic review, global survey, and Trend Analysis’, The Lancet Infectious Diseases, 19(2), pp. 143–155. doi:10.1016/s1473-3099(18)30647-9.
- Liu, Y. et al. (2012) ‘Identification of a novel HIV type 1 circulating recombinant form (CRF52_01B) in Southeast Asia’, AIDS Research and Human Retroviruses, 28(10), pp. 1357–1361. doi:10.1089/aid.2011.0376.
- Marcelino, J.M. et al. (2006) ‘Use of a new dual-antigen enzyme-linked immunosorbent assay to detect and characterize the human antibody response to the human immunodeficiency virus type 2 envelope GP125 and GP36 glycoproteins’, Journal of Clinical Microbiology, 44(2), pp. 607–611. doi:10.1128/jcm.44.2.607-611.2006.
- Nyamweya, S. et al. (2013) ‘Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis’, Reviews in Medical Virology, 23(4), pp. 221–240. doi:10.1002/rmv.1739.
- Pugach, P. et al. (2015) ‘A native-like sosip.664 trimer based on an HIV-1 subtype B env gene’, Journal of Virology, 89(6), pp. 3380–3395. doi:10.1128/jvi.03473-14.
- Sanders, R.W. et al. (2013) ‘A next-generation cleaved, soluble HIV-1 env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies’, PLoS Pathogens, 9(9). doi:10.1371/journal.ppat.1003618.
- Valadés-Alcaraz, A., Reinosa, R. and Holguín, Á. (2022) ‘HIV transmembrane glycoprotein conserved domains and genetic markers across HIV-1 and HIV-2 variants’, Frontiers in Microbiology, 13. doi:10.3389/fmicb.2022.855232.
- Vallari, A. et al. (2011) ‘Confirmation of putative HIV-1 Group P in Cameroon’, Journal of Virology, 85(3), pp. 1403–1407. doi:10.1128/jvi.02005-10.
- Wang, Q., Finzi, A. and Sodroski, J. (2020) ‘The conformational states of the HIV-1 envelope glycoproteins’, Trends in Microbiology, 28(8), pp. 655–667. doi:10.1016/j.tim.2020.03.007.