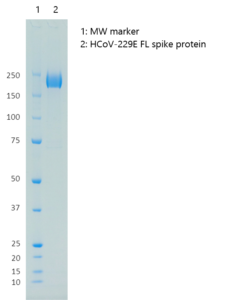
Human Coronavirus 229E Spike Glycoprotein (Full-Length), Sheep Fc-Tag (HEK293)
$943.93 – $3,585.38 excl. VAT
Human coronavirus 229E spike glycoprotein is a recombinant HCoV-229E full-length spike protein, with sheep Fc-tag, manufactured in HEK293 cells, with HCoV-229E spike protein soluble domain has been C-terminally tagged with a sheep Fc-tag. Amino acids 533-540 have been replaced with 9 AA glycine-serine linker to prevent protein cleavage by host cell proteases.
HUMAN CORONAVIRUS 229E SPIKE GLYCOPROTEIN (FULL-LENGTH), SHEEP FC-TAG (HEK293)
Human coronavirus 229E spike glycoprotein is a recombinant HCoV-229E full-length spike protein, with sheep Fc-tag, manufactured in HEK293 cells, with HCoV-229E spike protein soluble domain has been C-terminally tagged with a sheep Fc-tag. Amino acids 533-540 have been replaced with 9 AA glycine-serine linker to prevent protein cleavage by host cell proteases.
PRODUCT DETAILS – HUMAN CORONAVIRUS 229E SPIKE GLYCOPROTEIN (FULL-LENGTH)
-
- Human coronavirus 229E full-length spike glycoprotein (NCBI: QEG03785.1, Strain/Isolate: HCoV_229E/Seattle/USA/SC0865/2019).
- Protein contains a C-terminal sheep Fc-tag with apparent molecular weight of 200 kDa.
- Protein was produced in HEK293 cells and purified from culture supernatant by Protein G affinity chromatography.
- Presented as liquid, Dulbecco’s phosphate buffered saline pH7.4, sterile filtered.
BACKGROUND
Coronaviruses are enveloped, positive-stranded RNA viruses with a genome of approximately 30 kb (Lai, 1990). In animals they are significant veterinary pathogens, often causing severe disease (Holmes, 2001). Prior to early 2000, only two human coronaviruses were recognized, Human coronavirus 229E (HCoV-229E) and Human coronavirus OC43 (HCoV-OC43) which are a common cause of upper respiratory tract infections (Chibo and Birch, 2006). In late 2002, a third human coronavirus (SARS-CoV) was implicated as the aetiological agent of severe acute respiratory syndrome (SARS) (Drosten et al., 2003). Since then, several more human coronaviruses have been identified, including HCoV-NL63 associated with upper and lower respiratory tract infections (van der Hoek et al., 2004) and HCoV-HKU1 in patients with pneumonia (Woo et al., 2005) and Middle East respiratory syndrome (MERS). HCoV-229E was first described in 1966 when it was isolated from cell cultures inoculated with samples from diseased student volunteers (Hamre & Procknow, 1966).
Sequences related to HCoV-229E have been detected in captive alpacas and dromedary camels which may form an intermediate host for the virus. However, the viruses natural host is likely to be hipposiderid bats in Africa and these populations show much greater viral diversity than those from camelids. The spike genes of all HCoV-229E and camelid-associated 229E viruses contain deletions as compared to bat-associated viruses, which may have played a role in host switching. All 229E viruses from camels and all strains from bats also contain an additional gene (ORF 8) downstream of the nucleocapsid gene. Nucleocapsid proteins (nucleoproteins) are phosphoproteins that are capable of binding to helix and have flexible structure of viral genomic RNA. Nucleoprotein plays an important role in virion structure, replication and transcription of coronaviruses, as it localizes in both the replication/ transcriptional region of the coronaviruses and the ERGIC region where the virus is collected. It packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP) and plays a fundamental role during virion assembly through its interactions with the viral genome and membrane protein M. It also plays an important role in enhancing the efficiency of subgenomic viral RNA transcription as well as viral replication (Corman et al, 2018).
REFERENCES
- Chibo and Birch (2006). Analysis of human coronavirus 229E spike and nucleoprotein genes demonstrates genetic drift between chronologically distinct strains. Journal of General Virology, 87, 1203–1208.
- Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and Sources of Endemic Human Coronaviruses. Adv Virus Res. 2018;100:163-188.
- Drosten et al. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med348, 1967–1976.
- Holmes, K. V. (2001). Coronaviruses. In Fields Virology, 3rd edn, pp. 1187–1203. Edited by B. N. Fields, D. M. Knipe & P. M. Howley. Philadelphia: Lippincott Williams & Wilkins.
- Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121(1):190-193.
- Lai, M. M. (1990). Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol44, 303–333.
- Woo et al. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol79, 884–895.