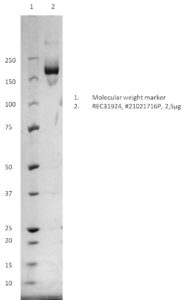
SDS-PAGE: Coomassie-stained reducing SDS-PAGE showing purified spike protein.
SARS-CoV-2 (B.1.1.7) Stabilized Spike Glycoprotein (Full-Length), His-Strep-Tag (HEK293)
$989.55 – $3,763.85 excl. VAT
UNITED KINGDOM (ALPHA) VARIANT
SARS-CoV-2, previously known as the 2019 Novel Coronavirus (2019-nCoV), causes the pandemic COVID-19 disease. Lineage B.1.1.7 (Variant of Concern 202012/01) is correlated with a significant increase in the rate of COVID-19 infection in the UK and includes amino acid substitutions 69-70del, Y144del, N501Y, A570D, P681H, T716I, S982A, D1118H relative to the prototypic Wuhan-Hu-1 strain.
SARS-COV-2 (B.1.1.7) STABILIZED SPIKE GLYCOPROTEIN (FULL-LENGTH), HIS-STREP-TAG (HEK293)
SARS-CoV-2 B.1.1.7 stabilized Spike glycoprotein (UK variant). SARS-CoV-2, previously known as the 2019 Novel Coronavirus (2019-nCoV), causes the pandemic COVID-19 disease.
PRODUCT DETAILS – SARS-COV-2 (B.1.1.7) STABILIZED SPIKE GLYCOPROTEIN (FULL-LENGTH), HIS-STREP-TAG (HEK293)
- SARS-CoV-2 B.1.1.7 full-length spike glycoprotein (“UK variant”).
- SARS-CoV-2 HexaPro stabilized Spike glycoprotein containing deletions and amino acid changes in accordance with variant B.1.1.7 (“UK variant”): 69-70del, Y144del, N501Y, A570D, P681H, T716I, S982A, D1118H relative to Wuhan-Hu-1.
- This protein contains 6 amino acid changes to proline and a C-terminal T4 fibritin trimerization domain to stabilize the protein in its pre-fusion conformation as described in Hsieh, Goldsmith et al. 2020.
- Expressed in HEK293 and purified by affinity chromatography.
- Presented in DPBS at
BACKGROUND
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus that causes coronavirus disease 2019 (COVID-19). The sequence WIV04/2019, belonging to the GISAID S clade / PANGOLIN A lineage / Nextstrain 19B clade, is believed to be the original sequence infecting humans (Zhukova et al., 2020). However, there are many thousands of variants of SARS-CoV-2 (Koyama et al., 2020) and subtypes of the virus can be placed into much larger groupings such as lineages or clades.
Lineage B.1.1.7 (Variant of Concern 202012/01) is correlated with a significant increase in the rate of COVID-19 infection in the UK. The two earliest sampled genomes were collected on 20-Sept-2020 in Kent and another on 21-Sept-2020 from Greater London. It was previously known as the first Variant Under Investigation in December 2020 (VUI – 202012/01)[20] and also as lineage B.1.1.7 or 20I/501Y.V1 (formerly 20B/501Y.V1). Genomes belonging to lineage B.1.1.7 form a monophyletic clade that is characterized by a large number of lineage-defining mutations. The B.1.1.7 lineage carries a larger than usual number of virus genetic changes with 17 mutations (14 replacements and 3 deletions) (COG-UK, 2020; Rambout et al., 2020). It is thought that the unusual genetic divergence of lineage B.1.1.7 may have resulted, at least in part, from virus evolution within a chronically infected individual.
B.1.1.7 mutations include spike position 501, one of the key contact residues in the receptor binding domain (RBD), and experimental data suggests mutation N501Y can increase ACE2 receptor affinity (Starr et al. 2020). N501Y has been associated with increased infectivity and virulence in a mouse model (Gu et al. 2020). P681H is a replacement of one of 4 residues comprising the insertion that creates a furin cleavage site between S1 and S2 in spike. The S1/S2 furin cleavage site of SARS-CoV-2 is not found in closely related coronaviruses and has been shown to promote entry into respiratory epithelial cells and transmission in animal models (Hoffmann et al., 2020; Peacock et al. 2020; Zhu et al. 2020). The spike deletion 69-70del has also occurred a number of times in association with other RBD changes (COG-UK, 2020).
REFERENCES
- COG-UK update on SARS-CoV-2 Spike mutations of special interest. Report 1. COG-UK, 19th December 2020.
- Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ, Wang Y, Teng Y, Zhao Z, Cui Y, Li Y, Li XF, Li J, Zhang NN, Yang X, Chen S, Guo Y, Zhao G, Wang X, Luo DY, Wang H, Yang X, Li Y, Han G, He Y, Zhou X, Geng S, Sheng X, Jiang S, Sun S, Qin CF, Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020 Sep 25;369(6511):1603-1607.
- Hoffmann M, Kleine-Weber H, Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol Cell. 2020 May 21;78(4):779-784.
- Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020 Jul 1;98(7):495-504.
- Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. 2020.
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.
- Zhukova A, Blassel L, Lemoine F, Morel M, Voznica J, Gascuel O. Origin, evolution and global spread of SARS-CoV-2. C R Biol. 2020 Nov 24. doi: 10.5802/crbiol.29.