In this blog, we explain how seasonal and pandemic influenza vaccines are developed, manufactured and validated, and introduce our contract services for the rapid development of custom influenza reagents.
The Influenza Viruses: A Quick Recap
The influenza viruses are single-stranded RNA viruses belonging to the Orthomyxoviridae family, typically 80-120nm in diameter and spherical or filamentous in shape. Influenza’s genome comprises eight single-stranded RNAs encased in a protein capsid, surrounded by a lipid bilayer from which the Haemagglutinin (HA), Neuraminidase (NA) and Matrix (M1, M2) proteins protrude. The Orthomyxoviridae family comprises four major types of influenza virus (A–D), which are classified by their NA and M1 proteins; the most clinically relevant are the co-circulating influenza A and B viruses. For influenza type A, at least 16 highly variable HAs (H1-18) and 9 distinct NAs (N1-11) have been recognised so far [1]; the influenza B virus has a similar viral structure to type A, but lacks H/N subtypes.
Influenza virus infections (known as the flu) occur in distinct outbreaks of varying severity every year, with periods of peak incidence (known as flu seasons) oscillating between the northern and southern hemispheres on a seasonal basis [2]. Influenza infections usually affect the upper respiratory tract, with mild symptoms, including fever, headache and malaise. However, even a run-of-the-mill flu can be severe in otherwise healthy people, and lethal to those who are elderly or frail, with complications including pneumonia, organ failure and secondary infections. Combined, seasonal influenza is thought to cause an estimated one billion infections and 290,000-650,000 deaths, worldwide each year [3].
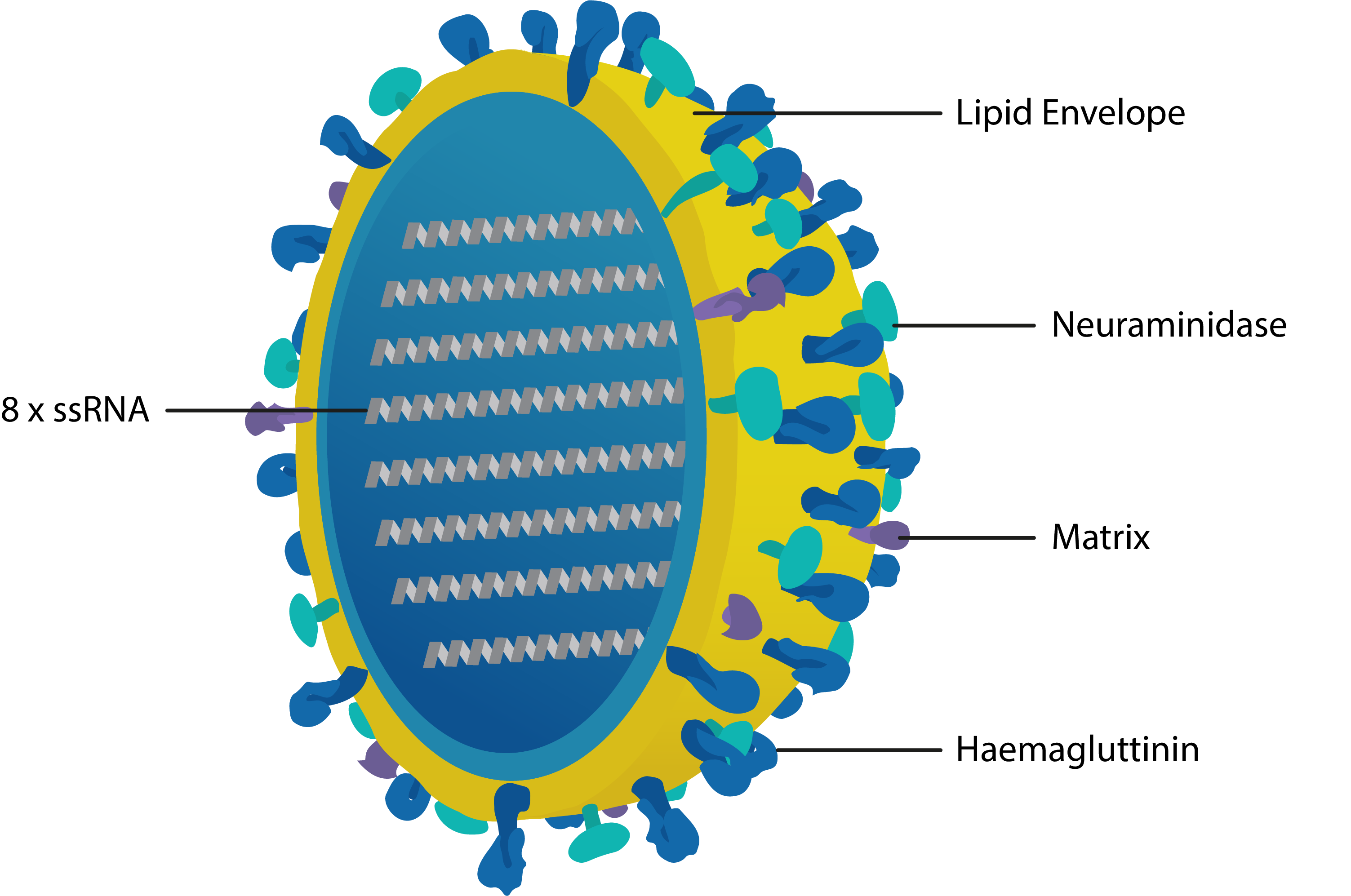
Cross-section of influenza virus with key proteins labelled. Haemagglutinin recognises cell-surface receptors and mediates cell entry; Neuraminidase cleaves neuraminic (sialic) acid residues to promote the release of viral progeny; Matrix 1 mediates assembly of the membrane complex and confers structural integrity; Matrix 2 is a selective proton channel that regulates viral pH; Nucleoprotein (not shown) binds the 8 strands of ssRNA and mediates packing, trafficking, transcription and replication.
The influenza viruses are arguably one of the most successful endemic and pandemic human pathogens. This can largely be attributed to their host diversity and ability to mutate. Beyond being RNA viruses with an innate ability to quickly generate genetic variants, influenza viruses are especially adept at undergoing significant genomic changes to evade recognition by adaptive immune responses. Influenza has achieved this to great effect through a potent combination of antigenic drift and shift: antigenic drift is a continuous process in which the development of adaptive immunity by hosts creates selection pressures for mutations in influenza’s Haemagglutinin proteins [4]. As HA contains various structurally tolerant regions and influenza RNA polymerase lacks proofreading activity, HA mutations can quickly accumulate. This allows HA’s epitopes to quickly change and escape host recognition, without hindering influenza’s ability to gain entry to cells.
On the other hand, antigenic shift is characterised by abrupt changes to HA and NA through genome reassortment with other influenza subtypes that occurs when two different influenza viruses co-infect the same cell. This can result in highly advantageous gain-of-function mutations that confer the virus with new disease characteristics or host tropisms. While all influenza viruses undergo antigenic drift, only the influenza A viruses are able to undergo antigenic shift due to their extensive animal reservoirs, which provide the viral diversity to facilitate such events. By shifting the epitopes of their surface antigens so drastically, influenza A strains are able to catch the immune system completely off-guard and rapidly proliferate through human populations. Indeed, the most devastating human influenza epidemics, including the 1918 Spanish Flu, the 1968 Hong Kong flu, the 2004 Bird Flu and the 2009 Swine Flu, were all the result of human/non-human influenza A strains that had recombined [5].
Vaccinating Against a Moving Target
To prevent both seasonal and pandemic influenza, various countermeasures are employed, including community alerts, antiviral medication, personal protective equipment and screening. However, undoubtedly the most effective means of controlling influenza is with the widespread use of prophylactic vaccines, which not only reduce transmission, but limit the severity of infection, particularly in risk groups such as children or the elderly. Design strategies for the development of influenza vaccines are based on the understanding that HA’s head – and especially its loop regions – is the most potent target for neutralising antibody responses, given their exposure and role in pathogenesis. However, considering that these immunogenic loops are also the least conserved regions of HA, in addition to the potential emergence of new HA subtypes via recombination, newly developed vaccines are perpetually threatened by rapid obsolescence.
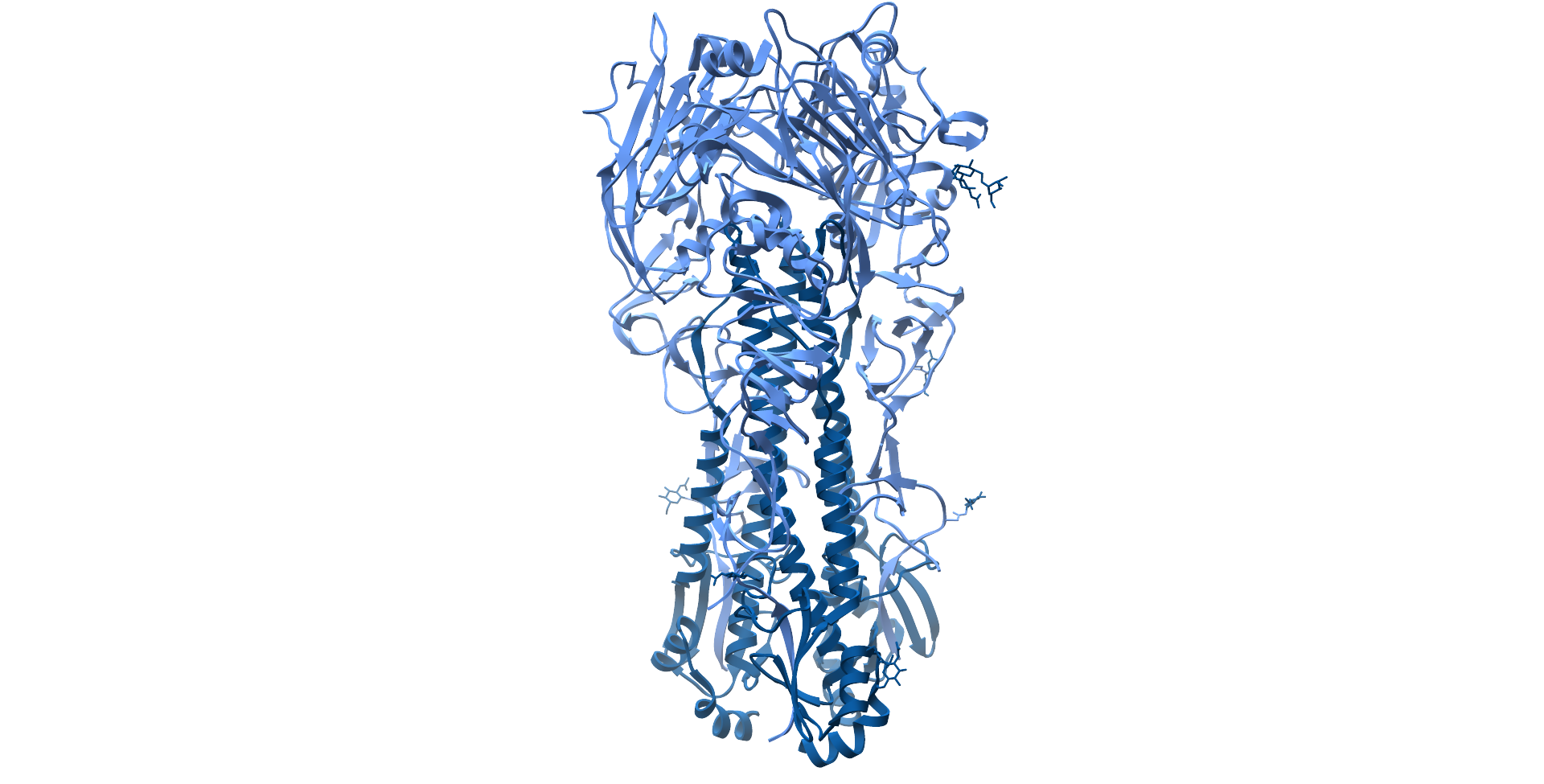
Influenza A H1N1 Haemagglutinin protein ectodomain, with hyerpvariable head region coloured light blue and stalk region coloured dark blue (PDB: 4GXX).
Because of this, new influenza vaccines must be formulated on a biannual basis for upcoming flu seasons in the Northern and Southern hemispheres. Selection of these formulations is led by the World Health Organisation (WHO), whose Global Influenza Surveillance and Response System (GISRS) of around 150 global laboratories monitor the spread of influenza and aim to predict the emergence of new strains. Collectively, these laboratories, known as National Influenza Centres (NICs), test over two million respiratory specimens annually, performing primary virus isolation and preliminary antigenic characterisation, as well as guiding the development and interpretation of tools to anticipate both seasonal and emergent zoonotic strains. Once isolates are taken from confirmed patients, they are then shipped to specialist WHO labs, known as WHO Collaborating Centres (CCs), for higher level antigenic and genetic analysis – the result of which forms the basis of recommendations for vaccine composition [6].
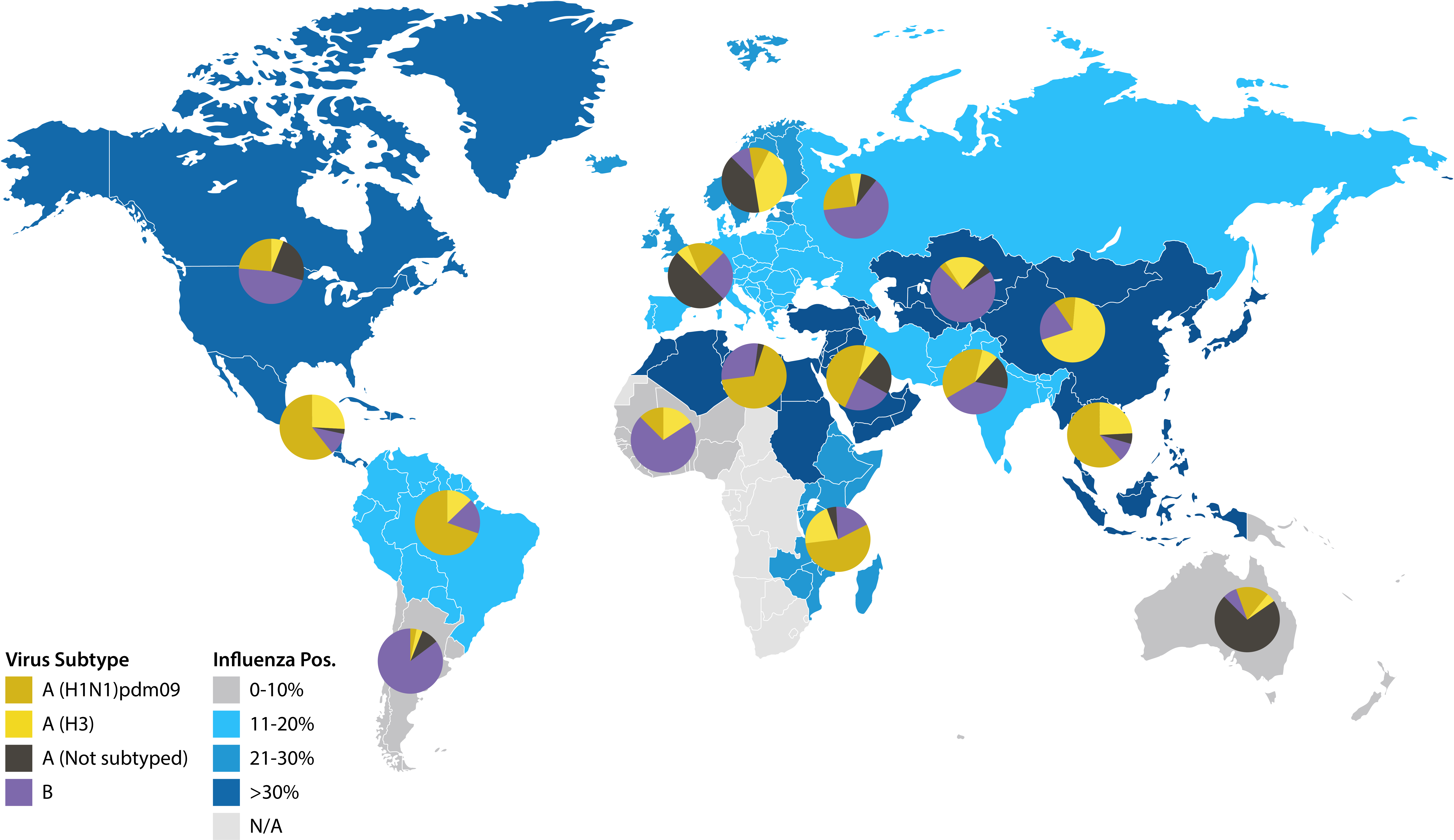
Map illustrating January 2020 influenza surveillance data from NICs. Reproduced from the World Health Organization, Influenza update 359 [7].
After 135 flu seasons and 3 pandemics, GISRS continues to represent a model platform for global collaboration in public health. Year-round surveillance of seasonal influenza forms not only the bedrock of seasonal vaccines, but facilitates responses to emerging pandemics, as early warning helps to mitigate the consequences of outbreaks and maintain the efficacy of existing vaccines. However, predicting future transmission is no easy feat, and is based on well-informed, but nonetheless, fallible guesswork. As a result, seasonal formulations don’t always match the predominating strain, resulting in only a small degree of cross-protection and markedly lower efficacy [8, 9]. The 2014/15 flu season, for example, caused a global spike in excess deaths that were largely attributed to differences between the vaccine and circulating H3N2 strains that resulted in only 19% vaccine effectiveness [10].
The second major caveat of influenza vaccine development is the slow and resource-costly manufacturing processes required to produce vaccines at scale. Most inactivated (IIV) and attenuated influenza vaccines (AIV) are manufactured using millions of fertilised chicken eggs, with separate strains being harvested and combined to produce trivalent or quadrivalent formulations. Standard doses are then filled into containers for vials, syringes or sprays for administration. As this process typically takes 6–8 months end-to-end, most vaccines are semi-obsolete by the time they are made publicly available [5]. For pandemic vaccines, it’s a similar story: upon detection of a major outbreak, a scramble ensues as multiple manufacturers race to produce candidates. The drawbacks of manufacturing parallel those of seasonal vaccines, and were evidenced by the devastating 2009 H1N1 swine flu epidemic, in which delayed vaccine administration was estimated to have caused hundreds of thousands of additional deaths [11, 12].
To overcome these limitations, various recombinant, next-generation vaccine platforms are being developed with more conserved epitopes that expand on the strain-specific nature of existing formulations. Specifically, platforms are being developed to protect against all strains within a subtype (e.g. all H1 strains) and multiple subtypes (e.g. H1/H5/H9), with the ultimate goal of creating a truly ‘universal’ pan-influenza A/B vaccine that can elicit lifelong immunity [13]. The benefits of such a vaccine are evident: as influenza continues to be the only human disease that requires annual vaccination, it is estimated that replacing just 10% of seasonal vaccines with a universal vaccine would avert 6,300 influenza-related deaths and save one billion US dollars in direct healthcare costs per year in the United States alone [14].
However, the development of a universal vaccine has not been without its challenges. As of yet, the breadth of cross-protection conferred by existing vaccine candidates is limited to small groups of subtypes and don’t stimulate truly heterotypic protection [15]. Furthermore, considering that the immunity conferred by seasonal vaccines wanes after about 6 months [16], it is not yet clear how universal vaccines will confer long-lasting protection. Consequently, traditional influenza vaccines are likely to stay for the foreseeable future, with a continued requirement for surveillance and prediction efforts by the GISRS.
The Validation Bottleneck
Given the strict timelines necessary for development, manufacture and distribution, influenza vaccines are subject to unique regulatory processes that require close communication between public health authorities, vaccine manufacturers and regulatory agencies.
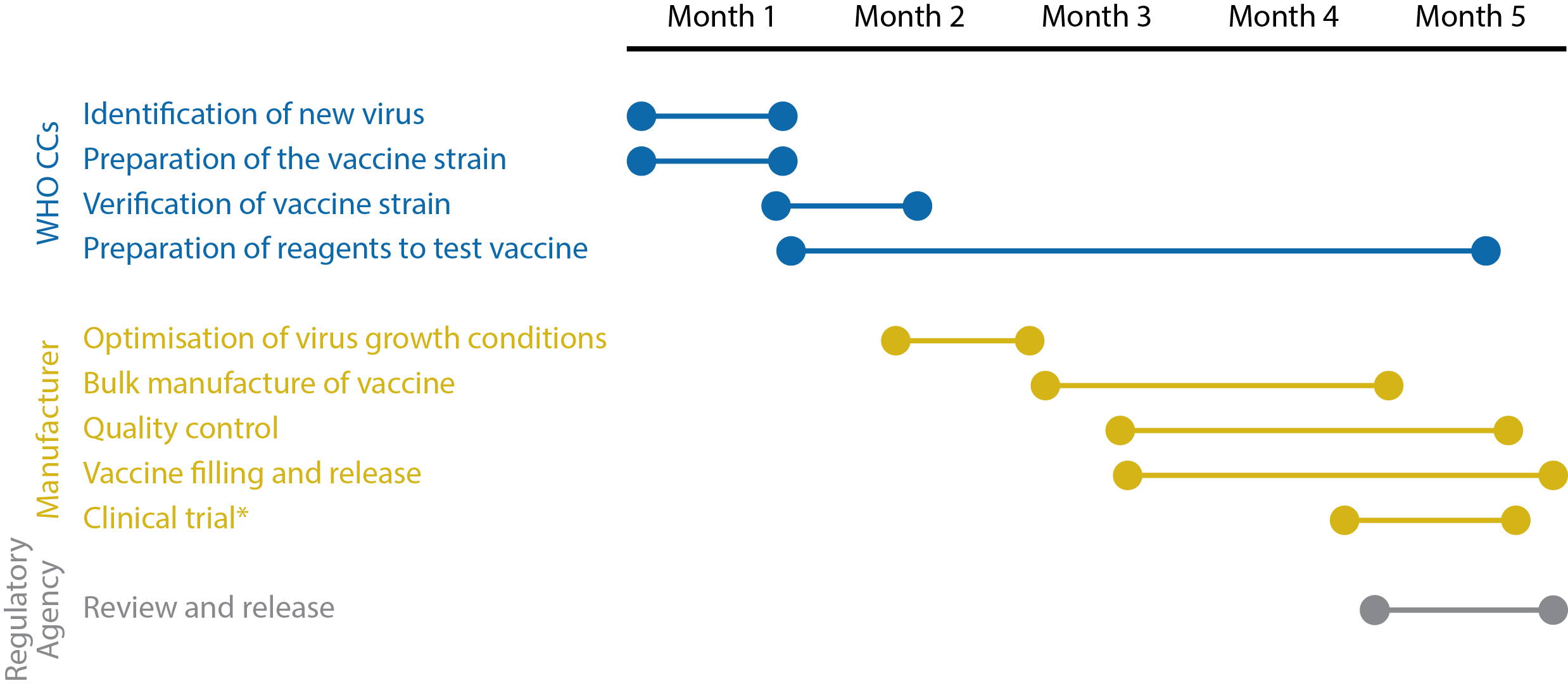
Timeline illustrating the key stages of influenza vaccine development. *Clinical trials are not carried out in all countries. Adapted from the World Health Organization’s Pandemic influenza vaccine manufacturing process and timeline [17].
A key stage in the development pipeline is the validation of vaccines, which are assessed for their ability to induce specific antibody responses in animals and humans. For decades, this was performed using Haemagglutinin inhibition assays (HAIs) and single radial haemolysis assays (SRHs) to determine whether vaccinated sera met strict, pre-defined standards. However, recent studies have shown that humans generate a remarkably broad immune response to influenza, and that upon subsequent challenge with novel strains, pre-existing influenza-specific B- and T-cell reactivity can have a positive or negative effect on an individual’s ability to neutralise the virus [18]. It therefore became apparent that HAI and SRH measurements were not necessarily the most informative and were poorly optimised for novel platforms, leading to a more diverse approach that allowed the use of a wider range of evidence-based endpoints [19].
Today, platforms such as enzyme-linked immunosorbent assays (ELISAs) [20] and antigen microarrays [21] are popular means of serological analysis, given their ability to qualitatively and quantitatively determine the humoural response to vaccination and compare it with natural infection. These tests are especially useful for assessing responses in those at the extremes of age that cannot be included in comparative efficacy studies, but for whom vaccines are most needed [22]. However, as influenza strains, and their subsequent vaccines vary season-to-season, appropriate serological tests are not readily available and must be developed for a specific formulation [23]. In addition, a broad range of reagents must be developed in parallel and calibrated for use as controls to ensure sufficient assay specificity. As such, the development of assays and the critical reagents used to design them can become major bottlenecks in the validation process. And although assay development does not typically delay vaccine availability, it is a yearly challenge to produce assays on time, with recent increases in the number of vaccine strains leading to growing concerns about the timely availability of reagents [24].
Contract Influenza Services Now Available
To support influenza vaccine development, The Native Antigen Company now offers custom contract services to rapidly develop panels of high quality influenza antigens using our proprietary VirtuE expression system, which ensures full glycosylation and proper folding. To demonstrate our capabilities, we have produced a panel of HA and NA proteins from multiple influenza strains, with a success rate of >90% of those attempted.
Complicating vaccine design, influenza viruses are able to utilise host-cell machinery to post-translationally add N-linked glycans to the antigenically-conserved domains of HA and shield them from immune recognition (known as glycan shielding) [25]. Therefore, the removal of glycan moieties allows the recognition of more immunogenic epitopes in vaccine formulations to increase the breadth and degree of protection.
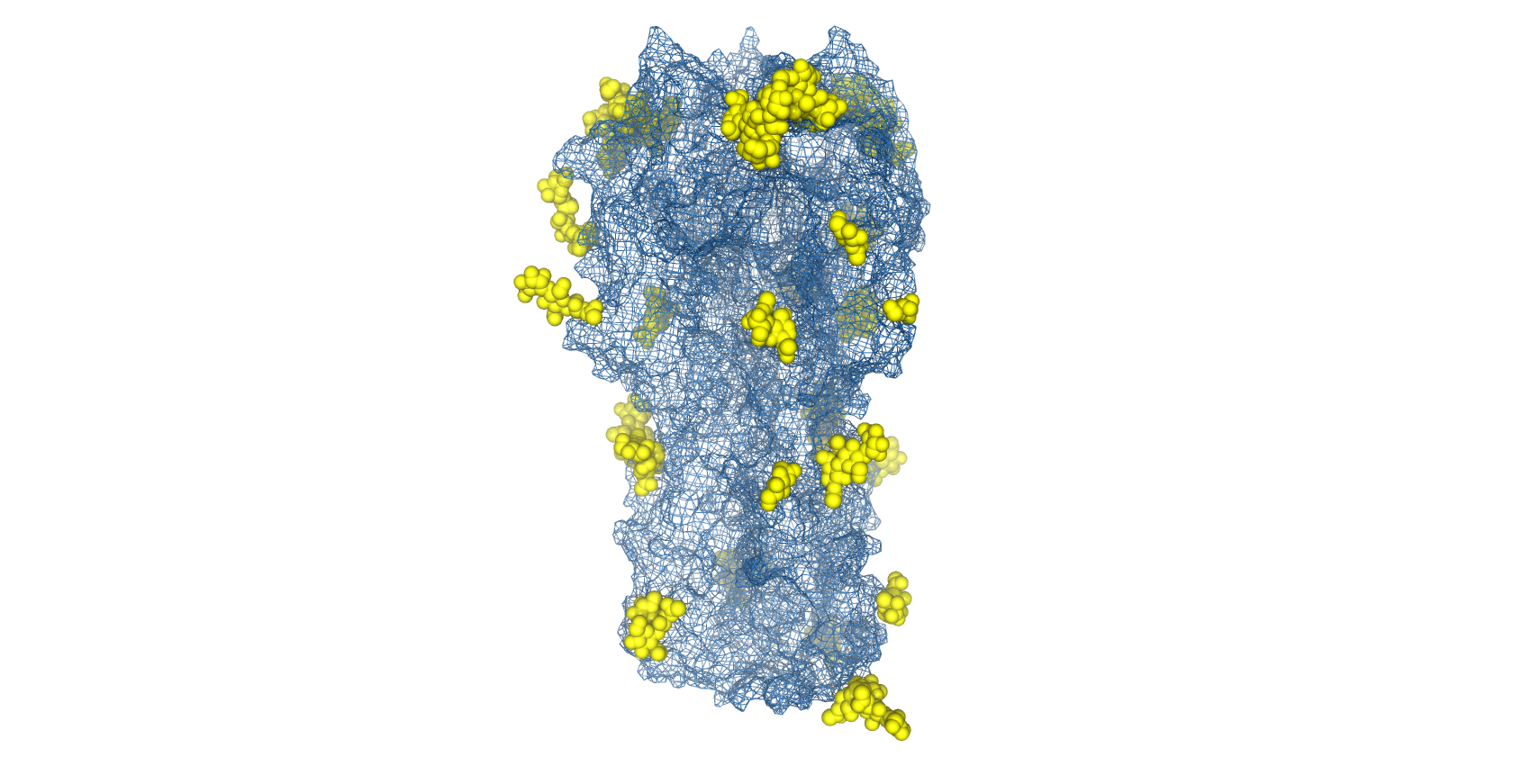
Structure of H1N1 Haemagglutinin with glycans shown as space-filled valences (yellow) and protein topography shown as mesh surface (blue) (PDB: 4UO2).
However, as glycans are necessary for protein folding and trimerisation, complete de-glycosylation by prokaryotic expression [26], treatment with tunicamycin [27] or PNGase F digestion [28] are not viable strategies. To ablate specific glycosylation sites, The Native Antigen Company offers the ability to carry out site-directed mutagenesis of specific asparagine residues, while still facilitating proper protein folding. In addition, we also offer antibody generation and immunoassay development services to provide further support to your influenza research.
For a bespoke proposal and consultation, please get in touch using the form below
References
1) https://www.cdc.gov/flu/about/viruses/types.htm
2) https://www.cdc.gov/flu/season/index.html
3) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5935243/
4) https://science.sciencemag.org/content/326/5953/734
5) https://www.sciencedirect.com/science/article/abs/pii/S128645791400327X?via%3Dihub
6) https://www.who.int/influenza/vaccines/virus/recommendations/en/
7) https://www.who.int/influenza/surveillance_monitoring/updates/2020_01_20_update_GIP_surveillance/en/
8) https://www.cdc.gov/flu/prevent/how-fluvaccine-made.htm
10) https://www.cdc.gov/flu/vaccines-work/effectiveness-studies.htm
11) https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(12)70121-4/fulltext
12) https://www.nejm.org/doi/10.1056/NEJMra1208802
13) https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-020-0626-6
14) https://pubmed.ncbi.nlm.nih.gov/31548402/
15) https://www.nature.com/articles/d41586-019-02751-w
16) https://pubmed.ncbi.nlm.nih.gov/28039340/
17) https://www.who.int/csr/disease/swineflu/notes/h1n1_vaccine_20090806/en/
18) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3667171/
20) https://pubmed.ncbi.nlm.nih.gov/29803776/
21) https://pubmed.ncbi.nlm.nih.gov/31403629/
22) https://onlinelibrary.wiley.com/doi/full/10.1111/irv.12383
23) https://pubmed.ncbi.nlm.nih.gov/28189147/
24) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5820418/
25) https://pubmed.ncbi.nlm.nih.gov/24638204/
26) https://pubmed.ncbi.nlm.nih.gov/19822741/
27) https://pubmed.ncbi.nlm.nih.gov/2738090/
28) https://pubmed.ncbi.nlm.nih.gov/24469815/