Through much of 2020, SARS-CoV-2 accumulated mutations at a steady, yet unspectacular rate. However, as global cases approached 100 million by the end of the year, multiple variants began to emerge. Exhibiting more considerable genomic changes, some variants have since shown to be more infectious and are able to escape the neutralising ability of certain monoclonal antibodies. In this blog, we discuss what is known about the new variants, highlight the role of neutralisation assays in characterising them, and introduce our new RBD/ACE2 Neutralisation Assay Development Kits.
Mutational Emergence
As SARS-CoV-2 emerged in late 2019, labs began to sequence a growing number of patient isolates. Within days, a Chinese-Australian group published the first SARS-CoV-2 draft genome sequence, opening the gates for a flood of research [1]. Scientists around the globe began to investigate COVID-19 biology, develop diagnostic tests and design candidate vaccines, marking what would become one of the research-intensive periods in human history. Aside from where the virus had actually originated, one of the biggest underlying questions was how, and to what extent, SARS-CoV-2 would mutate. To much relief, this was shown to be modest at best — no thanks to its proof-reading polymerase (Nsp14) shared with SARS- and MERS-CoV [2].
However, as the pandemic bloomed through the spring and summer, this virus was given an ever-increasing opportunity to mutate. Sequencing revealed an unusual burden of mutations occurring in the Spike gene, whose protein product is responsible for mediating cell-entry and subsequent pathogenesis. Of these, D614G (Asp614 → Gly) drew considerable attention following its rapid ascent to consensus. However, as the mutation was found to confer only mild improvements to viral infectivity and shedding, interest petered out and attention again returned to other facets of SARS-CoV-2 biology [3].
Nonetheless, as global cases continued to soar through late 2020, new mutations kept appearing. This came to a head in December as the world approached 100M cases, when three novel variants of concern (VoC) were all detected within the space of a month: B.1.1.7 in the UK [4], B.1.351 (501Y.V2) in South Africa [5] and B.1.1.24/P.1 in Brazil [6].
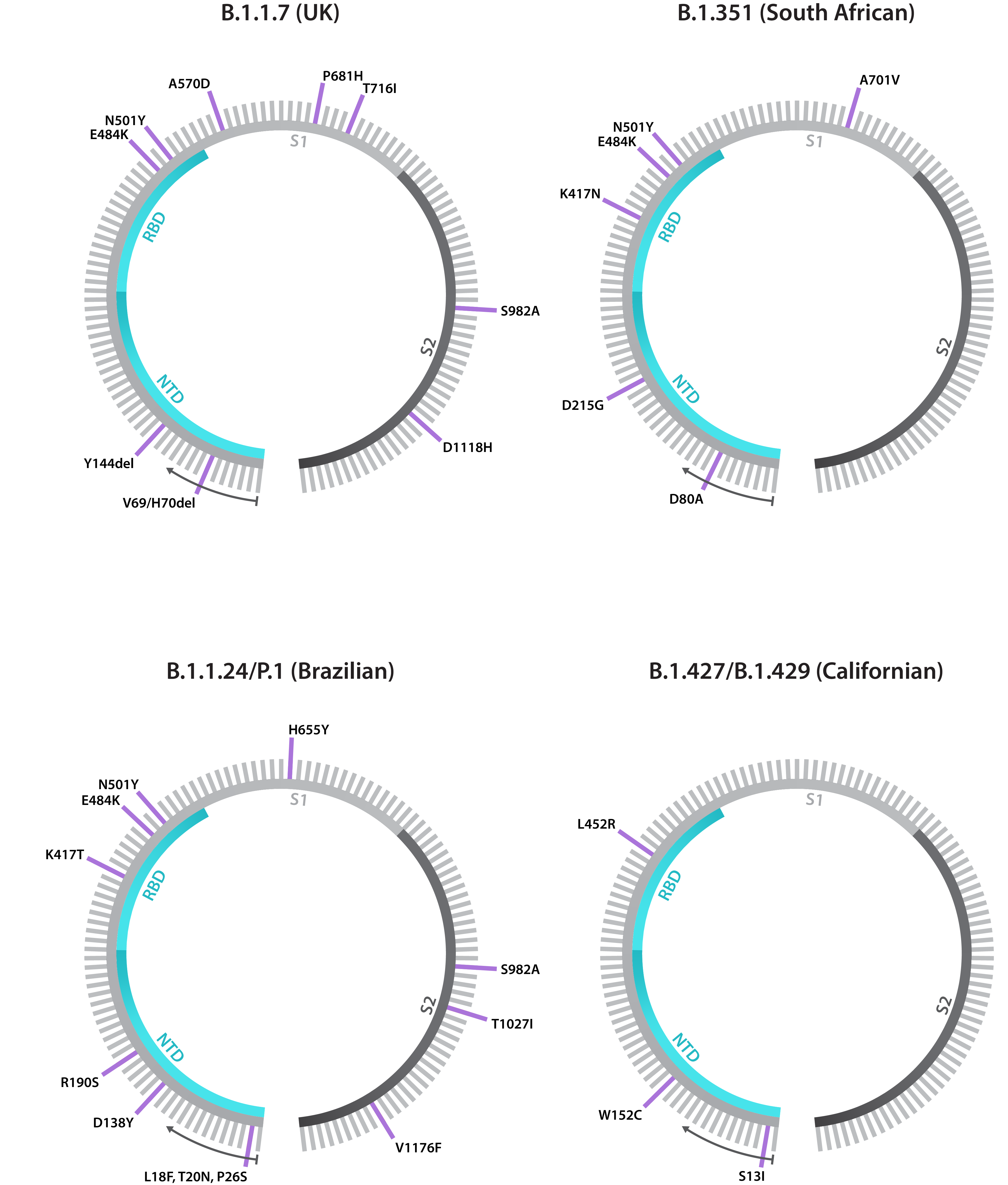
Schematics showing annotated Spike gene sequences for the B.1.1.7, B.1.351, B.1.1.24/P.1 and B.1.427/B.1.429 variants, including both point mutations and deletions. Note that each lineage has shown to exhibit Spike gene diversity, making these diagrams subject to change.
While D614G has shown that mutation is nothing new for the coronaviruses, what made the emergence of these variants so surprising was that each possessed a broad constellation of mutations not seen together before. Coinciding with the regional emergence of these variants, countries also witnessed surges in case numbers, soon ringing alarm bells amongst the global public health community [7]. Compelling enough, and combined with the low levels of population immunity, many countries have since to re-implemented lockdowns and travel restrictions.
Making Sense of the Mutations
For ongoing public health and vaccination programs, characterising variants has become a major priority. Multiple variants have already shown to be more transmissible, exacerbating global caseloads and disease severity. Moreover, there has been growing concern surrounding the fate of COVID-19 immunity, as all currently-available vaccines target the original Wuhan-Hu-1 strain’s Spike protein. These fears were stoked in early 2021, when in vitro data showed reduced sera neutralisation of variants and South African vaccine trials resulting in lower-than-expected protection, potentially due to the locally-circulating variant [8, 9]. Reinvigorating research efforts, many researchers have now pivoted their work to better characterise these variants and support the development of putative countermeasures.
The constellation of Spike mutations that has emerged is substantial and ever-changing, and different substitutions, deletions and combinations have showed the ability to confer a range of different traits, through an array of mechanisms. Those that have gained the most attention are K417N, E484K, and N501Y, all of which reside in Spike’s receptor-binding domain (RBD) — the immunodominant region responsible for ACE2 binding and the major target of neutralising antibodies [10]. All three have emerged repeatedly and independently of one another, suggesting they are advantageous low hanging fruit for the virus to adopt, alongside a string of other, less significant mutations. These mutations and others have shown to out-compete prototypic sequences, confer higher ACE2 affinity, and increase viral loads [11, 12, 13]. E484K, for example, has shown to confer up to 10-fold greater resistance to convalescent patient and vaccinee sera [14], and evade binding of multiple RBD-specific monoclonal antibodies [15].
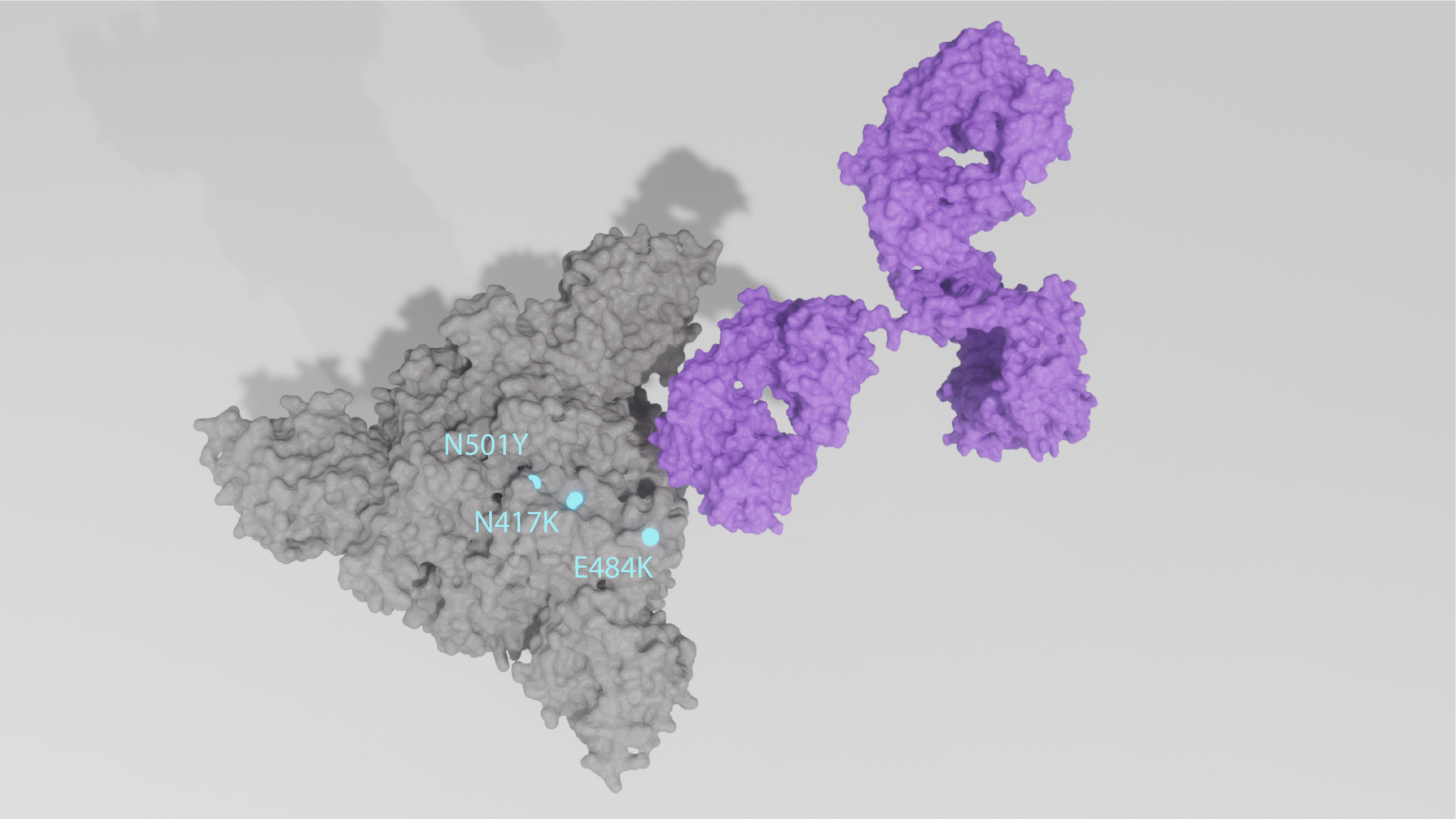
X-ray crystal structures of mutant Spike trimer in closed conformation being bound by IgG antibody. Key mutations are highlighted cyan and labelled accordingly.
In addition, Spike has also shown a tendency to lose small portions of its N-terminal domain (NTD) entirely. The H69/V70 deletion found in the B.1.1.7 (UK) variant, for example, has shown to contribute to increased infectivity and PCR target failure in diagnosis [16]. And finally, while recombination hasn’t gained significant interest outside of SARS-CoV-2’s putative origins and spillover, evidence suggests that it may be occuring and contributing to the emergence of novel variants [17]. Recombination events through co-infecting variants, in particular, may facilitate more rapid and drastic changes to the SARS-CoV-2 genome.
Neutralising or Not?
The emergence of multiple variants in recent months is a reminder that the pandemic could still result in some last-minute surprises. As the majority of vaccines provoke an immune response to multiple Spike epitopes, the breadth of antibodies elicited from vaccination are expected to continue conferring at least a moderate degree of protective efficacy and significantly reduce the incidence of severe disease [18, 19]. However, the proportion of vaccine coverage needed to achieve herd immunity is not yet clear, and the bar may rise higher and higher if new variants show increased transmissibility in naïve and immune individuals. This in turn, may be exacerbated by immune-mediated selection pressures from therapeutic-, patient- or vaccine-induced antibodies that accelerate antigenic drift [20].
Coming as a clear wake-up call, a concerted global effort is now underway to track, characterise and contain these novel mutations. In this regard, consensus and intrahost genome sequencing continues to play a fundamental role in surveillance [21], generating a wealth of epidemiological data that can indicate whether novel variants are more infectious, immune-resistant or virulent. Prospective studies using deep mutational scanning and predictive computational simulations have also provided early indications of mutation functionality [22, 23]. However, as we still lack a means of comprehensively and systematically assessing immunity at the patient level, significant gaps remain in our scientific arsenal.
Neutralising antibody titres correlate well with the protective efficacy of vaccines, and so have largely defined correlates of protection [15]. However, most large-scale serological studies to-date have relied on rapid tests, which lack the functionality or quantifiability needed to measure neutralising efficacy, and suffer from relatively low sensitivity and specificity [24]. To address this, a new generation of immunoassays are required to enable more standardised means of assessing variant sensitivity to neutralisation by monoclonal antibodies and vaccinee sera, as well as vaccine protection of animals against challenge with novel variants [25].
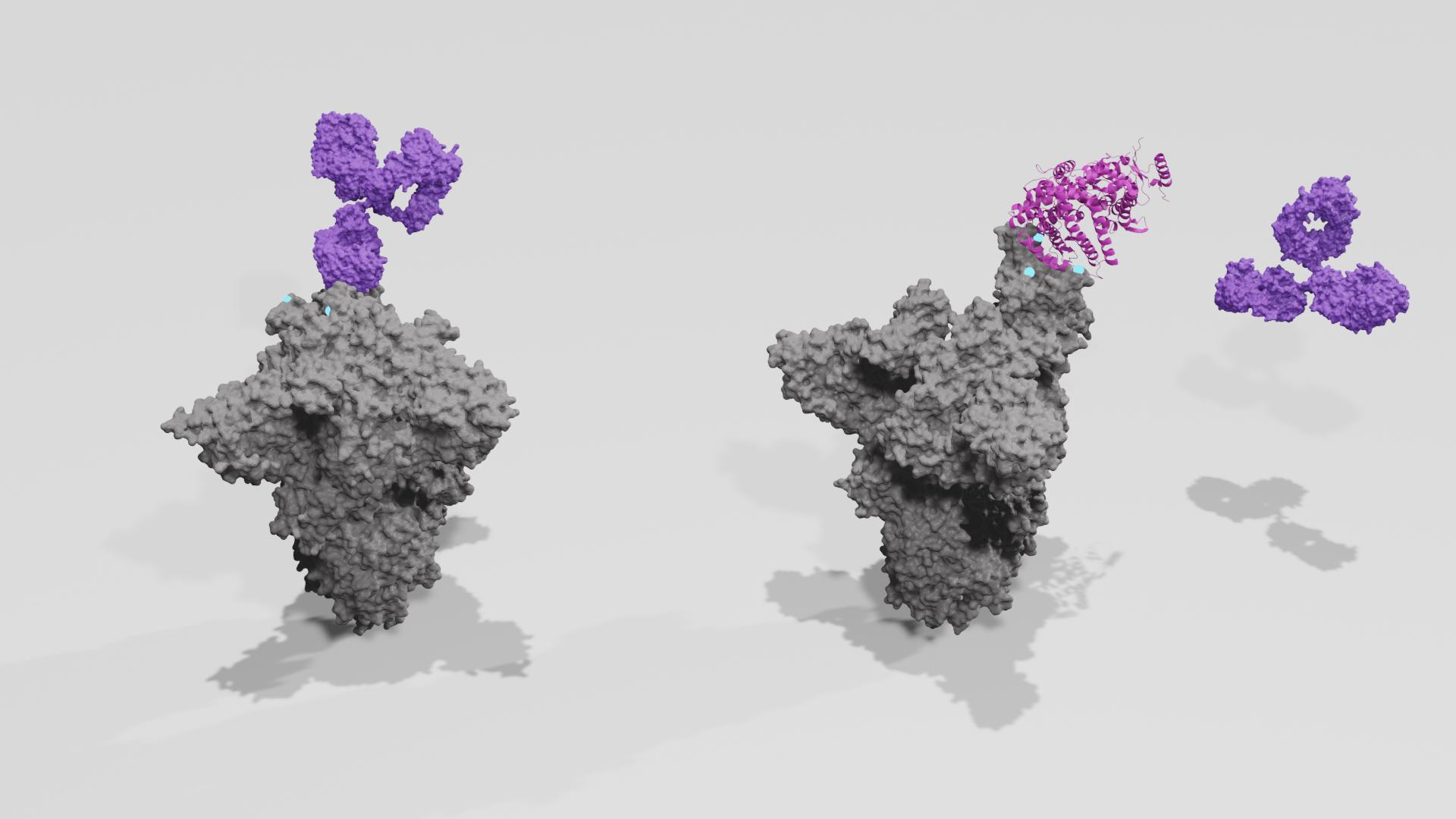
Above: Principles of competitive binding neutralisation assay. Above left: Antibody binds mutant Spike trimer; Above right: Antibody fails to neutralise Spike trimer, allowing ACE2 to bind RBD. Spike proteins are shown grey, point mutations are shown cyan, antibodies are shown purple, ACE2 is shown pink.
Alternative Approaches to Vaccine Design
It’s also worth remembering that SARS-CoV-2 is just one of seven known human coronaviruses, of which four have been circulating in human populations for decades causing mild forms of disease. While these endemic human coronaviruses (HCoVs) have largely been overlooked in favour of more clinically-relevant pathogens, they have gained significant attention since the emergence of SARS-CoV-2. Similar to the influenza viruses, HCoVs undergo continuous, albeit slower, antigenic drift, which combined with their weak induction of antibodies in humans, causes immunity to wane within a few years of infection [26, 27].
To no surprise, like SARS-CoV-2, HCoV Spike RBD’s show the highest levels of genetic drift, limiting patient cross-protection against different variants and species [28]. Incidentally, existing SARS-CoV-2 vaccine design strategies are based on the understanding that the RBD is the most potent target for neutralising antibody responses. However, as the RBD is the most poorly conserved Spike domain, monovalent formulations that rely on it risk becoming obselete.
As a result, the ever-shifting mutational landscape of SARS-CoV-2 and what we know about HCoV immunity has ignited new approaches to vaccine design strategies. Hoping to stay on step ahead, researchers are evaluating heterologous boosters, in which different Spike immunogens are used in secondary booster shots to broaden the patient’s immune response [29]. Taking a page out of influenza’s book, researchers have also begun to explore designs for universal vaccines that targets Spike’s more conserved regions to induce cross-protective antibody responses against a range of variants [30]. Chimeric Spike proteins, for example, could display a variety of different RBD variant epitopes, but retain the same sub-immunodominant regions within a single formulation [31].
However, striking the right balance between breadth and degree of protection is easier said than done, and developing such formulations by rational design would require a comprehensive understanding of vaccine-induced antibody profiles. Here, combinatorial evaluations of multiple existing and potential Spike mutations, and the functionality of the antibodies they elicit would be needed, This in turn, requires the development of bespoke neutralisation assays that can evaluate patient sera and be used in conjunction with serial passage.
Neutralisation Assay Development Kit Now Available
To support the investigation of SARS-CoV-2 variants and antibody functionality, The Native Antigen Company now offers a Neutralisation Assay Development Kit. This easy-to-use, cell-free kit contains all the key reagents required to measure SARS-CoV-2-antibody binding, including the Spike RBD of the prototypic Wuhan-Hu-1 strain, labelled ACE2, and positive and negative monoclonal antibody controls.
To see our Neutralisation Assay Development Kit and our in-house binding data, click the button below:
Our Spike RBD is produced with a full glycosylation pattern and proper folding to display native-like epitopes and minimise non-specific and/or cross-reactive antibody binding in assays. In addition to SARS-CoV-2 RBD, we offer a wide range of coronavirus reagents from a range of different expression systems. To see our full range, click the button below:
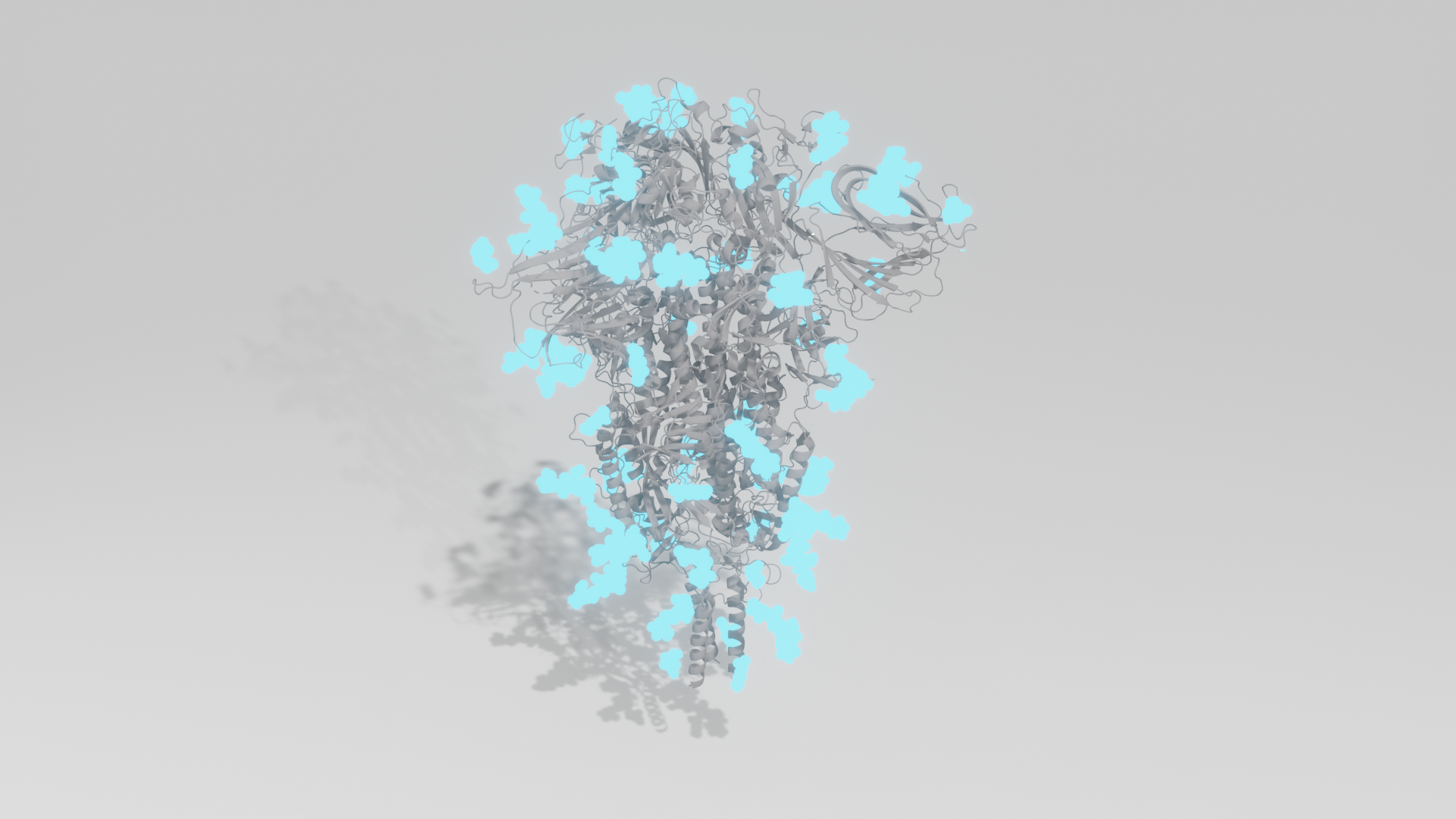
3D crystal structure of fully glycosylated SARS-CoV-2 Spike ectodomain trimer in closed confirmation. Protein is shown grey; glycans are shown cyan.
References
2) https://pubmed.ncbi.nlm.nih.gov/32853546/
3) https://www.sciencedirect.com/science/article/pii/S0092867420315373
6) https://www.medrxiv.org/content/10.1101/2020.12.21.20248640v1
7) https://www.sciencedirect.com/science/article/pii/S1567134820304779
10) https://www.nature.com/articles/s41586-021-03324-6
11) https://www.nature.com/articles/s41586-021-03426-1
12) https://www.bmj.com/content/372/bmj.n579
13) https://www.medrxiv.org/content/10.1101/2021.03.03.21252706v2
14) https://www.biorxiv.org/content/10.1101/2021.01.25.428137v3
15) https://www.nature.com/articles/s41586-021-03412-7
16) https://jamanetwork.com/journals/jama/fullarticle/2778599
17) https://www.biorxiv.org/content/10.1101/2021.01.21.427579v1
18) https://www.biorxiv.org/content/10.1101/2020.11.06.372037v1
19) https://pubmed.ncbi.nlm.nih.gov/33713594/
20) https://www.cell.com/cell-reports/fulltext/S2211-1247(21)00204-7
21) https://www.biorxiv.org/content/10.1101/2021.01.19.427330v1.full.pdf
22) https://www.sciencedirect.com/science/article/pii/S0092867420310035
23) https://www.sciencedirect.com/science/article/pii/S1931312820306247
24) https://www.nature.com/articles/s41467-021-22102-6
25) https://jamanetwork.com/journals/jama/fullarticle/2776542
26) https://pubmed.ncbi.nlm.nih.gov/2170159/
27) https://pubmed.ncbi.nlm.nih.gov/3008551/
28) https://jvi.asm.org/content/77/16/8801
29) https://www.tandfonline.com/doi/full/10.1080/22221751.2021.1902245
30) https://www.sciencedirect.com/science/article/abs/pii/S0092867420308126
31) https://jvi.asm.org/content/87/12/6542