While less-well known than some of their counterparts, the 229E, NL63, OC43, and HKU1 human coronaviruses are a significant cause of respiratory disease worldwide. The evolutionary histories and host associations of the endemic coronaviruses also provide important insights into the history of coronavirus emergence and the development of effective countermeasures. In this blog, we discuss the emergence of the endemic coronaviruses, what they may tell us about the epidemiology and immunology of SARS-CoV-2, and introduce our new range of HCoV cell lysates for the development of highly specific immunoassays.
Coronavirus Phylogeny and Emergence
The coronaviruses (CoVs) are a group of large, enveloped, positive-sense, ssRNA viruses belonging to the family Coronaviridae, responsible for respiratory, enteric and neurological forms of disease. All CoV genomes share a similar organisation, with the 5′ two thirds comprising the overlapping ORF1a and ORF1b reading frames that encode 16 nonstructural proteins, while the remaining portion of the genome includes reading frames for the Spike (S), Envelope (E), Membrane (M) and Nucleoprotein (N), in addition to various accessory proteins encoded by certain strains.
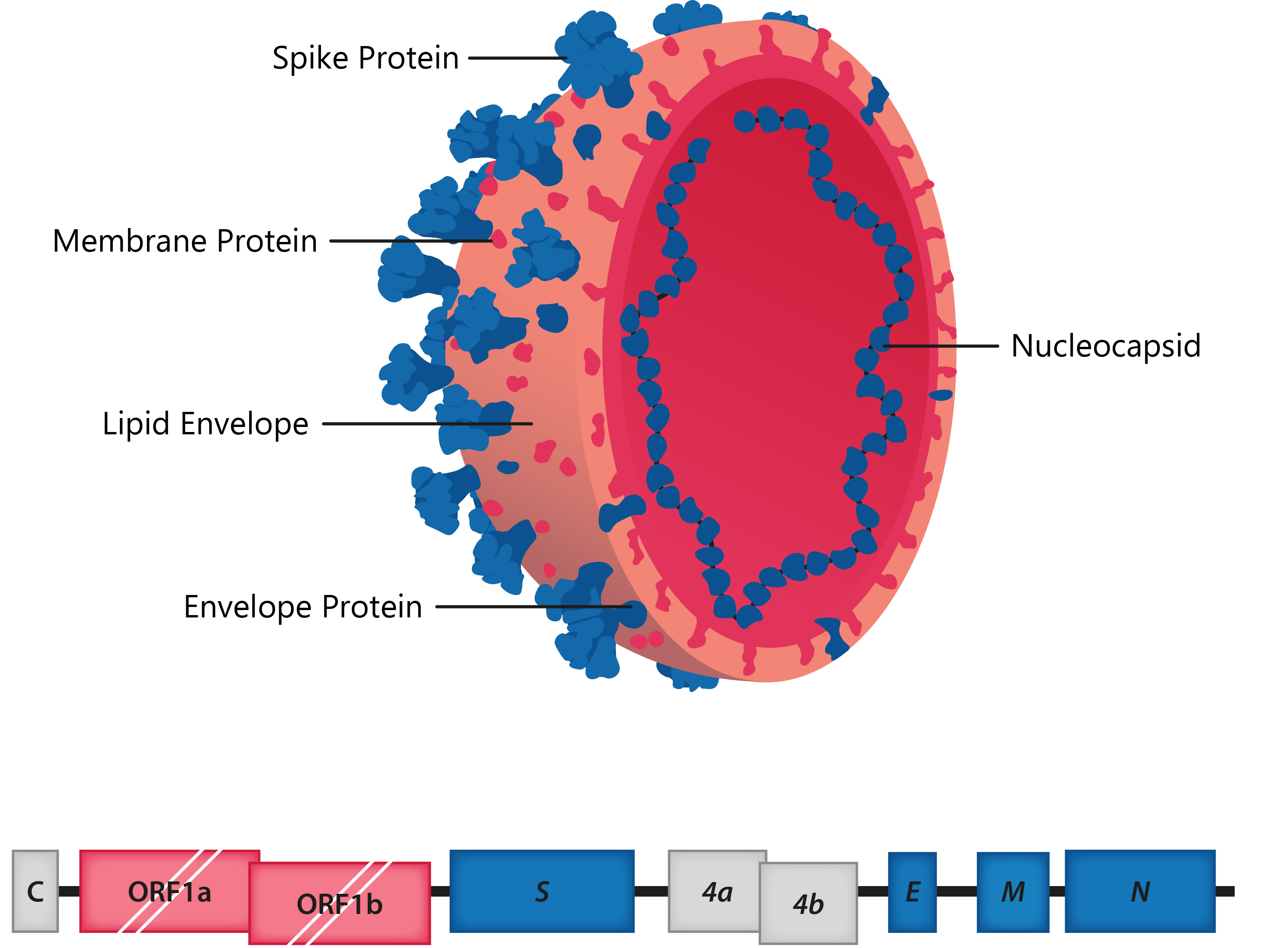
Above: Cross-section of coronavirus virion, with structural proteins labelled; Below: HCoV-229E genome structure.
The coronavirus family includes the subfamily Orthocoronavirinae, which is further divided into the alpha-, beta-, gamma- and deltacoronavirus genera. The alpha- and betacoronaviruses are known to infect mammals, such as humans, bats, pigs, camels and mice, whereas the gamma- and deltacoronaviruses are mostly avian, though they have also been isolated from whales, pigs and other mammals [1, 2].
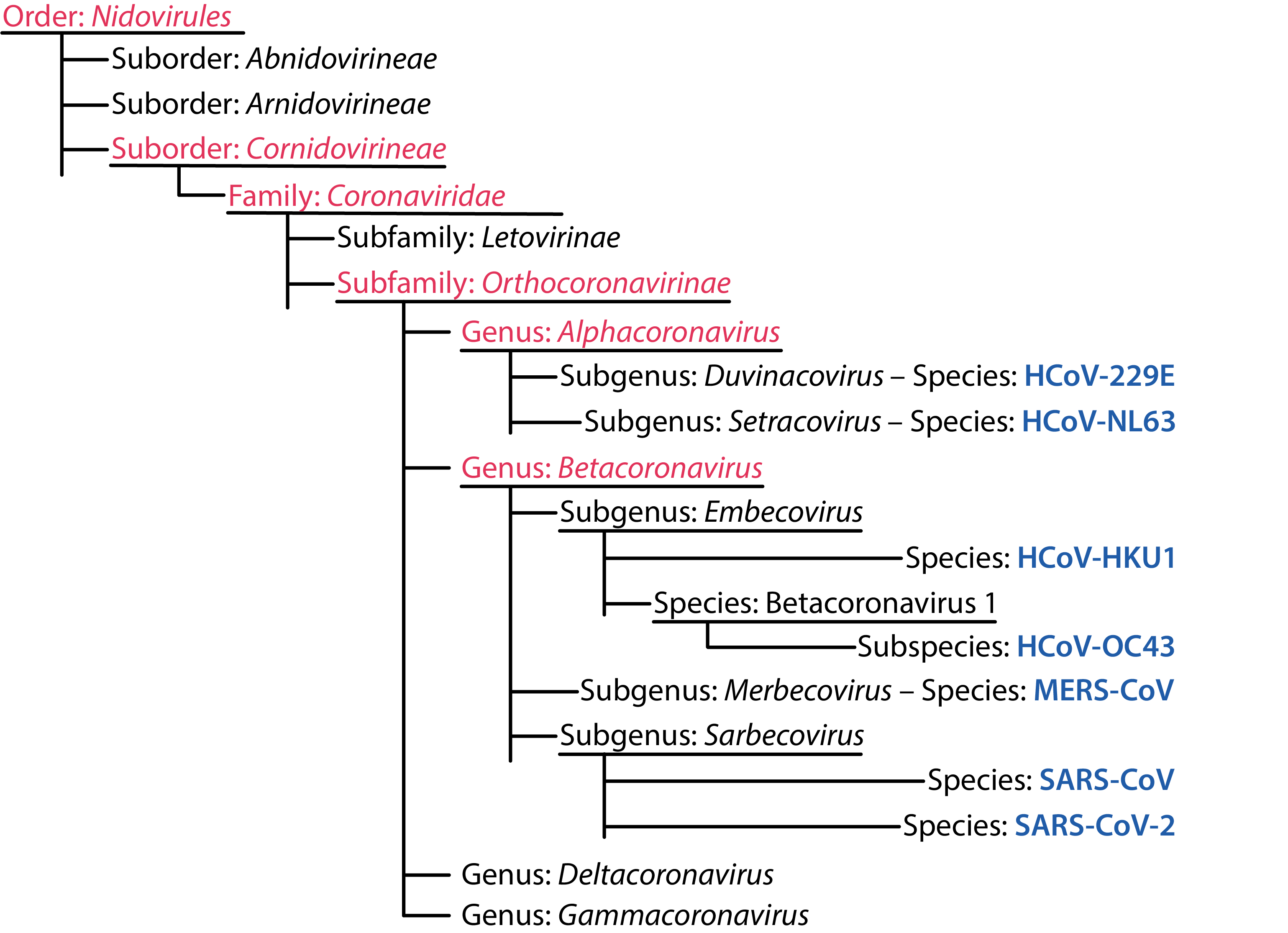
ICTV Human Coronavirus Taxonomy [3].
Following the emergence of multiple pathogenic strains in recent decades, CoVs have gained a reputation as highly effective spillover pathogens. The factors contributing to the rise in CoV emergence are not fully understood, though this phenomenon has been broadly attributed to two main factors:
+ Host Ecology
Coronaviruses can be found in a highly diverse range of different mammalian and bird host species. Combined with increasing industrialisation, farming, urbanisation, deforestation, transportation and climate change, this has provided a multiplicity of routes for coronaviruses to spillover into human populations. In particular, a large proportion of coronaviruses are thought to reside in bat reservoirs, which are particularly adept at facilitating cross-species transmission. Bats are an immensely diverse group, with over 1,000 species accounting for around a quarter of mammalian diversity [4], not to mention the range in climate, geography, social behaviours and movement patterns they experience. Likewise, bat CoVs show a great degree of genetic diversity and over 3,000 distinct bat CoVs are thought to exist [5], leading some to propose that most, if not all currently circulating coronaviruses may derive from bat zoonoses which may have established other animal reservoirs or intermediate hosts [6]. In addition to their role as vectors for disease, bats are also thought to augment virulence, given that a range of highly virulent viruses have emerged in recent decades (CoVs, Ebola and paramyxoviruses). While the causes of this phenomenon are not yet clear, it appears that bats have developed particularly strong anti-viral responses that allow them to tolerate a diversity of viral infections and create selection pressures for increased virulence in infecting strains, leading to more deadly spillovers in species that lack such adaptations [7].
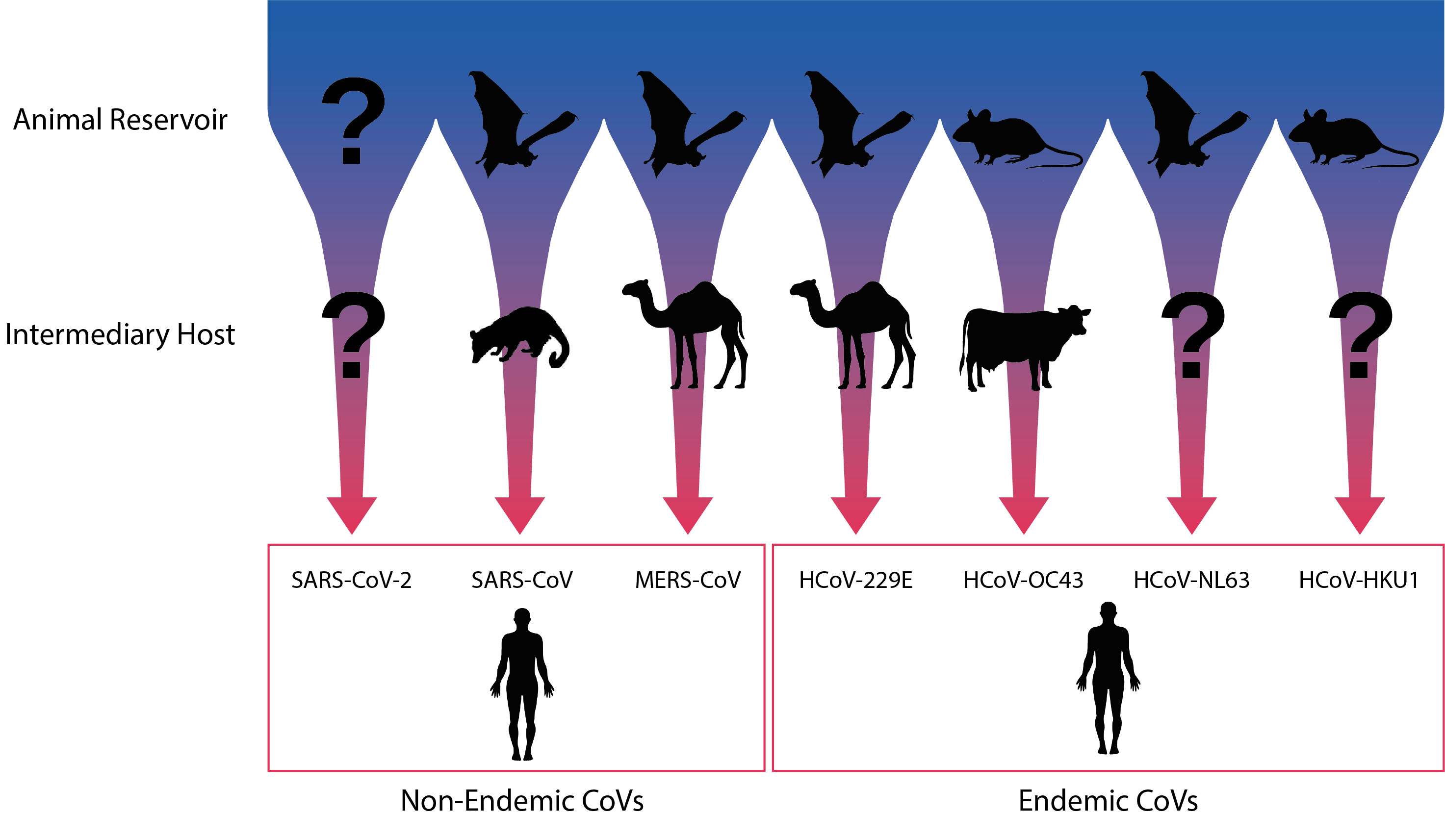
Diagram illustrating the natural and putative intermediate hosts of the seven human coronaviruses. Adapted from Corman et al.
+ Large and Mutable Genomes
In addition to their complex host ecologies, CoVs have exceptionally large genomes (26-32kb), in-fact the largest known of any RNA virus. Their genomes are also able to incur mutations at a high rate and incorporate various accessory proteins to facilitate rapid change [9, 10]. Moreover, the CoV replication cycle produces a series of subgenomic RNAs that can easily recombine with the genomes of closely related lineages by template switching [11]. In combination, these factors allow CoVs to quickly optimise their host interactions when transferring to a new species (e.g. changing receptor affinities) and undergo recombination to yield more radical changes (e.g. new tissue tropisms) that lead to enhanced pathogenesis and further spillover events.
The Endemic HCoVs & Their Antibody Responses
To date, seven coronaviruses have been identified in humans, of which SARS-CoV (2002), MERS-CoV (2012) and the newly-identified SARS-CoV-2 have caused significant epidemics with high mortality rates. Less well-known are the four remaining human CoVs: HCoV-229E, HCoV-OC43, HCoV-NL63 and HCoV-HKU1. Unlike the epidemic CoV strains, the HCoVs are established endemic pathogens found across much of the globe, causing milder forms of respiratory disease. Like other respiratory viruses, HCoVs also show a marked seasonality, with high detection frequencies in the winter months and are responsible for an estimated 15-30% of common colds [12]. But while the HCoVs have gained a reputation as relatively benign pathogens, they have been increasingly associated with a broadening range of clinical outcomes, including severe lower respiratory tract infections, neurological complications and death – especially when co-infecting with other respiratory pathogens [13]. The timeline below summarises the discovery and characteristics of the four HCoVs:
1966
HCoV-229E
The first human coronavirus was isolated in 1966 from cell culture by American scientists, Dorothy Hamre and John Procknow, naming it HCoV-229E after the student sample number from which it was taken [14]. HCoV-229E was classified as an alphacoronavirus and has since been associated with various respiratory symptoms, ranging from the common cold to pneumonia and bronchiolitis. Like other HCoVs, 229E infections are highly prevalent in children, with studies finding that most are infected in the first couple of years of life [15]. For the most part, 229E has had a fairly uneventful history. However, in 2007, a novel alphacoronavirus was identified following an outbreak of respiratory disease in alpacas in the US. Genomic studies revealed close relationships to 229E-related bat CoVs, placing it as an intermediate between the bat HCoV-229E-related viruses and HCoV-229E from humans [16, 17, 18, 19]. As camels are often kept alongside alpacas at animal trade fairs, it is thought that HCoV-229E originated in bats while camels and alpacas have served as a more recent zoonotic reservoir for human transmission.
1967
HCoV-OC43
Not long after the identification of 229E, HCoV-OC43 was isolated from human tracheal explants kept in organ culture (hence OC) in 1967 by researchers working at NIH [20]. The virus was found to be morphologically indistinguishable from 229E, but serologically distinct. OC43 was also found to differ significantly from the other HCoVs in that it belongs to the Embecovirus subgenus and includes an additional, shorter Spike protein called Haemagglutinin Esterase (HE) which is able to degrade specific sialic acid receptors. The virus has mostly been associated with mild upper respiratory tract infections, though it has also been suspected to play a role in more severe forms of neurological disease, including fatal encephalitis [21]. Phylogenetic analyses suggest that OC43 first emerged when a mouse-to-bovine CoV gained human tropisms near the turn of the 20th century following a gene deletion event that coincided with a respiratory pandemic [22].
2002
SARS-CoV
2004
HCoV-NL63
Following the emergence of SARS-CoV in 2002, the coronaviruses lost their mild-mannered reputation, leading many virologists to search for new strains. Researchers in the Netherlands soon reported the isolation of a novel coronavirus related to 229E in five people presenting with respiratory disease [23]. The virus – which came to be known as HCoV-NL63 (NetherLand 63) – is widespread and causes colds in most instances of infection. However, since its discovery, there have been hints that its effects may extend beyond the respiratory tract. In 2005, researchers found an association between NL63 and a Kawasaki-like disease characterised by the inflammation of blood vessels in primarily young children [24]. However, the findings were controversial with further studies finding no-such association [25]. NL63 likely emerged following a recombination event between NL63-like and 229E-like CoVs from Triaenops and Hopposideros bat species, respectively [26].
2005
HCoV-HKU1
Hot on the heels of NL63, was the isolation of a novel HCoV in an elderly patient with viral pneumonia at Hong Kong University (hence HCoV-HKU1) [27]. Follow-up studies by researchers around the world found HKU1 in circulation [28]. Most HKU1 infections have shown to be mild and self-limiting, with few deaths reported to date [29], however, HKU1 has also been associated with a high incidence of febrile seizures in children and a greater frequency of neurological sequelae than OC43, which it is often co-detected with, and with which it shares a mouse origin [30]. However, unlike OC43, HKU1 is most closely related to the mouse hepatitis virus (MHV) and is thought to have emerged slightly later in the early 1950s [22].
2012
MERS-CoV
2019
SARS-CoV-2
A defining characteristic of the HCoVs that has gained significant attention in recent months is their inability to induce lasting immune responses – a trait that likely developed to establish long-term persistence in human populations [31]. The cause of waning HCoV immunity is not entirely clear, though it has been attributed to a combination of insufficient immune stimulation from sub-clinical infections, poor cross-protectivity between serotypes and genetic drift [32]. Serosurveys have shown that the majority of individuals infected with HCoVs present with a baseline level of antibodies, with initial seroconversions tending to occur within the first few years of life [33]. However, after antibody titres reach a peak at 2 weeks post-infection, they gradually return to baseline levels 4-12 months later, where most individuals are again susceptible [34, 35]. Specific challenge studies have also been carried out with similar results. One study, for example, challenged patients with 229E followed by repeated challenge one year later [34]. In most cases, reinfections were successful, though with decreased symptom severity and shortened duration of shedding. In regards to cross-protection, the adaptive immune responses to CoVs are mainly directed towards the most variable regions of the structural proteins (e.g. Spike’s receptor-binding domain) that are not conserved across types [36]. Therefore, cross-protection between different HCoVs does not appear to play a significant role in immunity. The effects of antigenic drift, meanwhile, are poorly understood and further research is required to understand whether repeat infections are influenced by changing epitopes.
Implications for COVID-19 Immunity
As the SARS-CoV-2 pandemic has continued to unfold, there has been speculation that the immunology of the endemic HCoVs may foreshadow that of SARS-CoV-2. Early in the pandemic, many labs were detecting remnant RNA from the lungs of convalescent and recovered individuals, even when viable SARS-CoV-2 was not isolated, leading to false-positive results [37]. While such issues were swiftly resolved and PCR diagnosis of COVID-19 has since improved, emerging studies have again cast some doubt on the strength and duration of the COVID-19 immune response [38, 39, 40]. A study published in June, for example, compared the immune responses of asymptomatic and symptomatic COVID-19 patients, finding that 40% of asymptomatic individuals had undetectable levels of protective antibodies 2-3 months after infection, compared to 13% of symptomatic patients, along with lower levels of neutralising antibody [39]. At a glance, such findings would suggest HCoV-like immune longevity. However, it is too early to draw any conclusions and a major gap still lies in our knowledge of cell-mediated immunity. Longitudinal studies of patients that recovered from milder SARS infections, for example, also found antibodies decreasing after 2-3 years to minimal or non-existent levels [32], while memory T-cells have shown to persist for many years with high activity [41]. Yet given that SARS was contained relatively quickly, it is not known whether these memory T-cells would be sufficient to induce fast, protective responses [32].
Given the slow mutation rate of SARS-CoV-2, antigenic drift has yet to prove itself as a major factor, however, the role of cross-protection is less clear. A pre-print published in July, for example, showed that HCoV patient sera was able to neutralise live SARS-CoV-2 in the absence of SARS-CoV-2 antibody signals tested with ELISA [42], suggesting the potential for pre-existing HCoV antibodies to modulate the severity of SARS-CoV-2 infection. If this were the case, it would go some way in explaining the correlation between increasing age and susceptibility as HCoV infection rates (and therefore immunity) is higher in children than adults [43, 44]. However, cross-sectional population studies have cast doubt on HCoV/SARS-CoV-2 cross-protection, with little supporting evidence [45]. It is also worth considering that antibody-facilitated infection of phagocytic immune cells has been well characterised in SARS and MERS patients and is associated with similar aberrant inflammatory responses in COVID-19 infection [46].
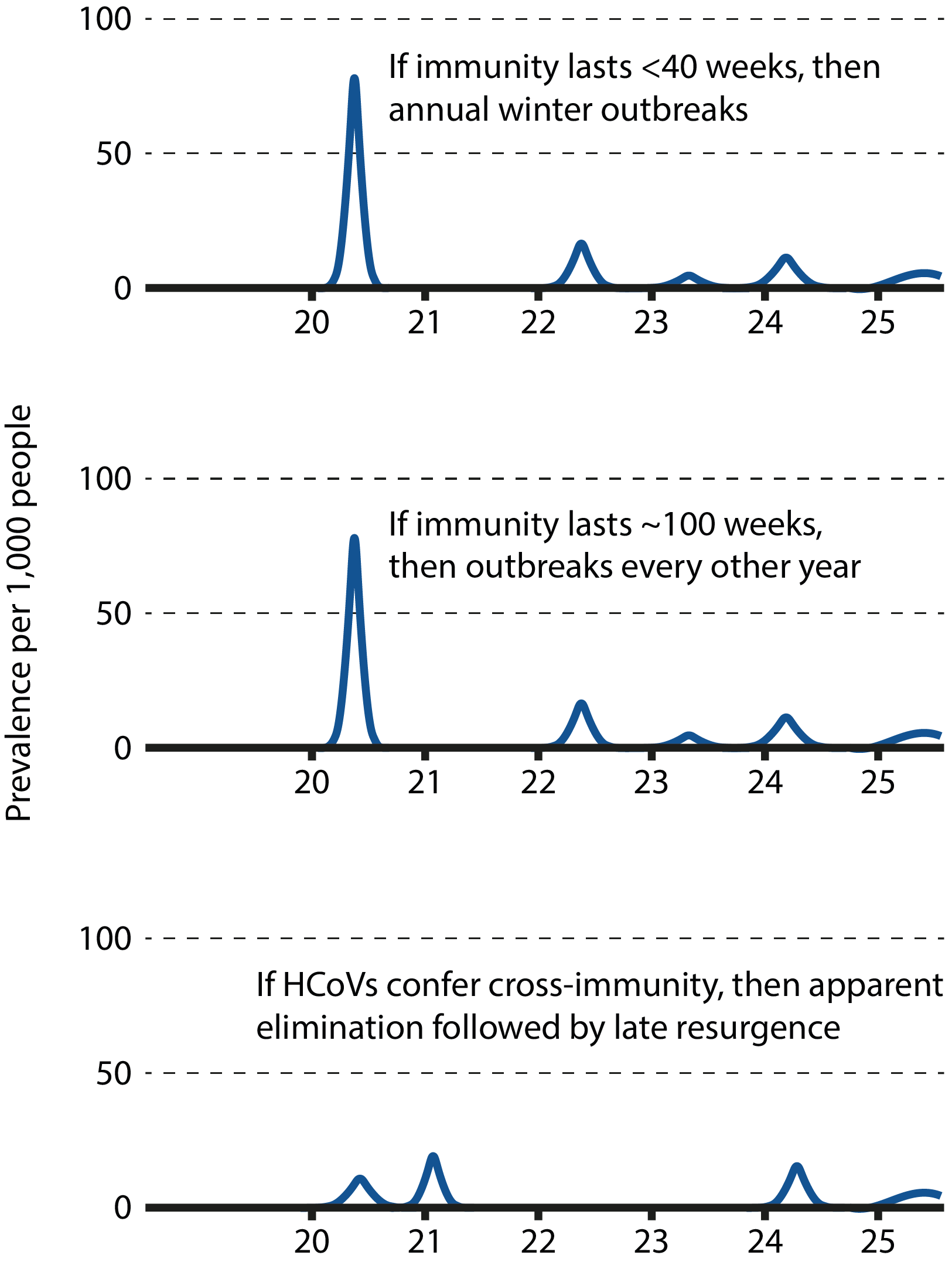
Graphs illustrating the different epidemiological scenarios according to the nature of human immunity to SARS-CoV-2. Adapted from Kissler et al.
The Role of Serology
While there is currently a lack of data describing the COVID-19 immune response, susceptibility to reinfection in the long-term remains a possibility. Modelling studies suggest that if immunity were short-term and could not be conferred by cross-protection, it would favour the establishment of long-term seasonal circulation with sporadic annual outbreaks [48]. However, if immunity were long-lasting, the virus could disappear for extended periods of time or even become eradicated from human and potential intermediary hosts. Distinguishing between these potential scenarios is key in formulating an effective, sustained public health response, with important consequences for both epidemiological modelling and the development of effective vaccines. In the case of epidemiological modelling, short-lived immunity would suggest the need for more long-term containment measures by public health authorities until effective countermeasures have been established; In the case of vaccines, researchers would need to consider measures to prolong vaccine-induced responses, such as dosage, boosters, or more potent antigenic determinants.
Regardless of whether SARS-CoV-2 reinfection is a possibility, distinguishing antibody functionality and titres between previous and de novo CoV infections is critical to understanding susceptibility, and would better define the correlates of protection for vaccines. Doing so requires large-scale, longitudinal serosurveys, supported by the development of high performance qualitative and quantitative immunoassays. Given the rate at which antibody titres have been shown to decline, these assays require especially high sensitivity, and in the case of vaccine diagnostics, should employ antigens heterologous to those used in formulations, such as Nucleoprotein.
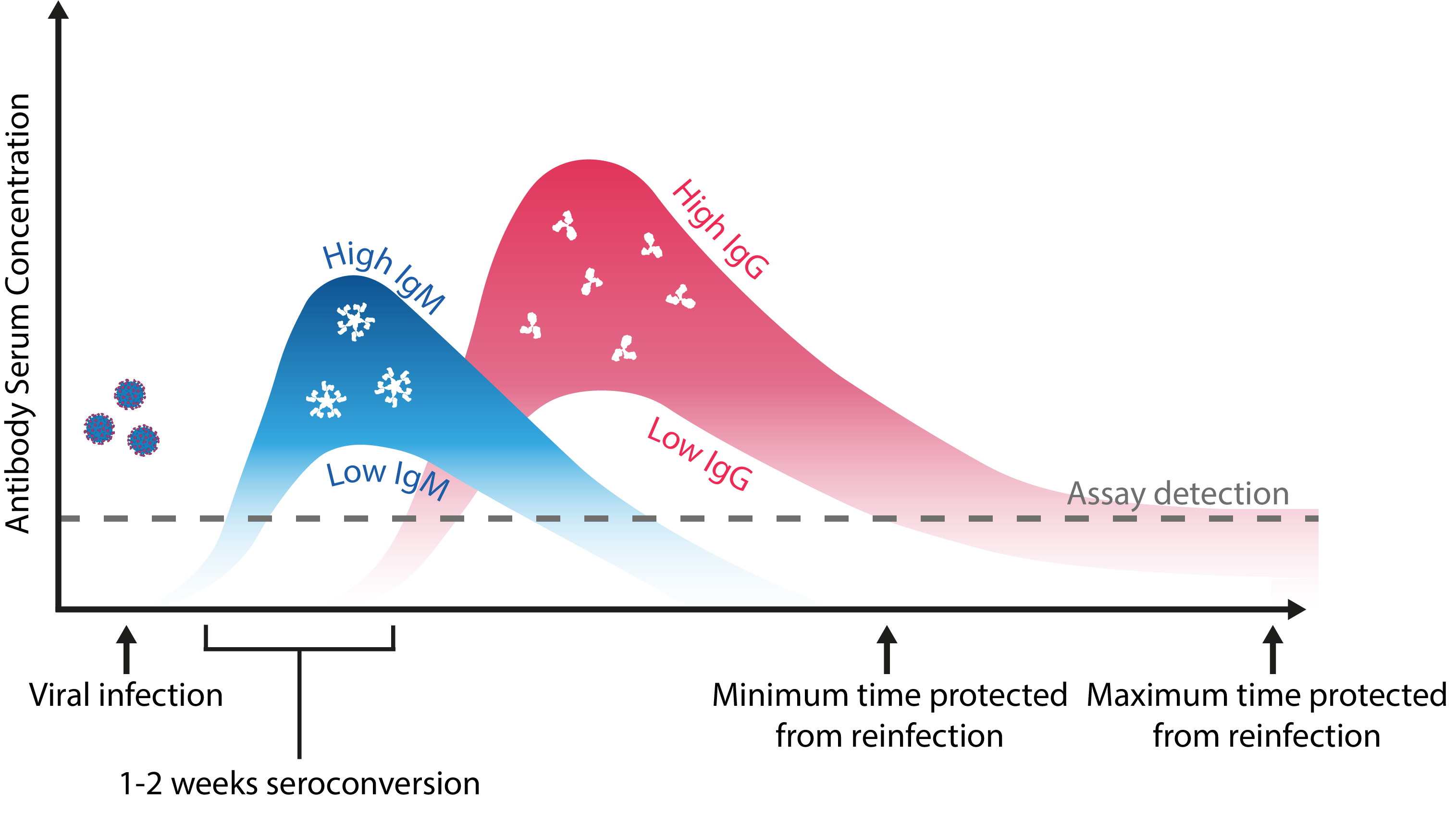
Schematic representation of the maximal and minimal limits of an antibody response to coronavirus infection. Adapted from Kellam and Barclay.
Finally, antibody diagnosis is further complicated by the potential of cross-reactive antibodies raised to HCoVs that can bind in immunoassays and produce false-positive results. Therefore, great care should be taken in assay validation studies to ensure that diagnostics are unable to detect HCoV-specific antibodies.
To support the ongoing research and development of coronavirus diagnostics, The Native Antigen Company now offers a range of highly purified HCoV lysates in addition to our HCoV Nucleoproteins, which can be used as controls for the development of highly specific immunoassays. For more information, please click on the links below:
We also offer an extensive and growing range of recombinant proteins for the non-endemic coronaviruses, including Spike proteins and various subunits, Nucleoproteins, Membrane/Envelope fusions and monoclonal antibodies.
References
1) https://jvi.asm.org/content/82/10/5084
2) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3302495/
4) https://www.sciencedirect.com/science/article/pii/S187962571830107X?via%3Dihub
5) https://academic.oup.com/ve/article/3/1/vex012/3866407
6) https://www.sciencedirect.com/science/article/pii/S0166354213003161
7) https://www.frontiersin.org/articles/10.3389/fimmu.2020.00026/full
8) https://www.sciencedirect.com/science/article/pii/S0065352718300010
10) https://journals.sagepub.com/doi/10.3181/0903-MR-94
11) https://www.pnas.org/content/107/27/12257
12) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7204879/
13) https://jcm.asm.org/content/48/8/2940
14) https://journals.sagepub.com/doi/10.3181/00379727-121-30734
15) https://pubmed.ncbi.nlm.nih.gov/18495857/
16) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2819850/
17) https://pubmed.ncbi.nlm.nih.gov/28077633/
18) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC224637/
19) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3528286/
20) https://pubmed.ncbi.nlm.nih.gov/5231356/
21) https://www.nejm.org/doi/full/10.1056/NEJMc1509458
22) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7111218/
23) https://www.nature.com/articles/nm1024
24) https://academic.oup.com/jid/article/191/4/499/937208
25) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2868826/
26) https://jvi.asm.org/content/91/5/e01953-16
27) https://jvi.asm.org/content/79/2/884.short
28) https://www.medscape.com/viewarticle/529443_1
29) https://pubmed.ncbi.nlm.nih.gov/16267760/
30) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1489438/
31) https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(04)01177-6/fulltext
32) https://www.microbiologyresearch.org/content/journal/jgv/10.1099/jgv.0.001439
33) https://pubmed.ncbi.nlm.nih.gov/24040960/
34) https://pubmed.ncbi.nlm.nih.gov/2170159/
35) https://pubmed.ncbi.nlm.nih.gov/3008551/
36) https://jvi.asm.org/content/77/16/8801
37) https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030
38) https://www.nejm.org/doi/10.1056/NEJMc2025179
39) https://www.nature.com/articles/s41591-020-0965-6
40) https://www.medrxiv.org/content/10.1101/2020.07.09.20148429v1
41) https://www.nature.com/articles/s41586-020-2550-z
42) https://www.biorxiv.org/content/10.1101/2020.05.14.095414v2
43) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7108278/
44) https://pubmed.ncbi.nlm.nih.gov/30241410/
45) https://www.medrxiv.org/content/10.1101/2020.06.29.20142596v1
46) https://www.sciencedirect.com/science/article/pii/S004268221400049X?via%3Dihub
47) https://pubmed.ncbi.nlm.nih.gov/32291278/
48) https://science.sciencemag.org/content/368/6493/860