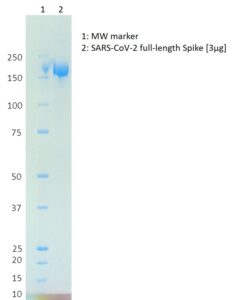
SDS-PAGE: Coomassie-stained SDS-PAGE of full-length SARS-CoV-2 spike protein.
SARS-CoV-2 Spike Glycoprotein (Full-Length), His-Tag (CHO)
$939.75 – $3,574.41 excl. VAT
Recombinant full-length SARS-CoV-2 spike glycoprotein, manufactured in Chinese hamster ovary (CHO) cells to high yield and purity. The furin cleavage site (amino acids 682-685) has been mutated from RRAR to SRAS and the transmembrane domain and intravirion part replaced with a glycine-serine linker and a His-tag. Horseradish peroxidase conjugated protein also available. SARS-CoV-2, previously known as the 2019 Novel Coronavirus (2019-nCoV), causes the pandemic COVID-19 disease.
SARS-COV-2 SPIKE GLYCOPROTEIN (FULL-LENGTH), HIS-TAG (CHO)
SARS-CoV-2 spike full-length (CHO) recombinant glycoprotein is manufactured in CHO cells to high yield and purity. The furin cleavage site (amino acids 682-685) has been mutated from RRAR to SRAS and the transmembrane domain and intravirion part replaced with a glycine-serine linker and a His-tag. Horseradish peroxidase conjugated protein also available. SARS-CoV-2, previously known as the 2019 Novel Coronavirus (2019-nCoV), causes the pandemic COVID-19 disease.
PRODUCT DETAILS – SARS-COV-2 SPIKE GLYCOPROTEIN (FULL-LENGTH)
- SARS-CoV-2 spike full-length (CHO) recombinant protein, manufactured in Chinese hamster ovary (CHO) cells.
- Contains Spike protein amino acids 1-1211. The furin cleavage site (amino acids 682-685) is mutated from RRAR to SRAS and the transmembrane domain and intravirion part replaced with a glycine-serine linker and a His-tag
- SARS-CoV-2 spike full-length (CHO) protein is purified by immobilized metal affinity chromatography.
- Presented in Dulbecco’s phosphate buffered saline (DPBS), pH 7.4.
- SARS-CoV-2 full-length spike binds ACE2 (REC31876) when used as capture antigen in immunoassay (ELISA), at a concentration between 0.5-5.0 µg/ml.
BACKGROUND
In late December, 2019, a number of patients with viral pneumonia (now called 2019-nCoV acute respiratory disease; COVID-19) were found to be epidemiologically associated with the Huanan seafood market in Wuhan, in the Hubei province of China. A novel, human-infecting coronavirus, provisionally named 2019 Novel Coronavirus (2019-nCoV) and since named SARS-CoV-2, was identified by genomic sequencing (Lu et al., 2020). SARS-CoV-2 is closely related (88% identity) to two bat-derived severe acute respiratory syndrome (SARS)-like coronaviruses, collected in 2018 in Zhoushan, eastern China, but were more distant from SARS-CoV (~79% identity) and MERS-CoV (~50% identity). However, although bats might be the original host of this virus, an animal sold at the seafood market in Wuhan may have acted as an intermediate host (Lu et al., 2020). Relative synonymous codon usage (RSCU) analysis suggests that SARS-CoV-2 is a recombinant between the bat coronavirus and an origin-unknown coronavirus, and it is thought that a Pangolin or other mammal is likely to have been the intermediate host. The recombination event occurred within the viral spike glycoprotein (Ji et al., 2020). Homology modelling shows that SARS-CoV-2 has a similar receptor-binding domain structure to that of SARS-CoV, despite amino acid variation at some key residues and is able to bind to the angiotensin-converting enzyme 2 (ACE2) receptor in humans (Lu et al., 2020). SARS-CoV-2 is a respiratory virus which spreads primarily through contact with an infected person through respiratory droplets generated when a person coughs or sneezes, or through droplets of saliva or discharge from the nose. The incubation period is believed to range from 2-11 days. Infection with SARS-CoV-2 can cause mild symptoms including a runny nose, sore throat, cough, and fever. However, it can be more severe for some people and can lead to pneumonia or breathing difficulties. The elderly, and people with pre-existing medical conditions (such as, diabetes and heart disease) appear to be more vulnerable to becoming severely ill with the virus (WHO, 2020).
The coronavirus spike (S) glycoprotein is a class I viral fusion protein on the outer envelope of the virion that plays a critical role in viral infection by recognizing host cell receptors and mediating fusion of the viral and cellular membranes (Li, 2016). SARS-CoV-2 entry into host cells is governed by the transmembrane spike (S) glycoprotein which forms homotrimers protruding from the viral surface. S contains an S1 subunit, responsible for binding to the host cell receptor, and a S2 subunit, facilitating fusion of the viral and cellular membranes. The S1 subunit comprises the receptor-binding domain(s) and contributes to stabilization of the prefusion state of the membrane-anchored S2 subunit, which contains the fusion apparatus. For other CoVs, S is cleaved at the boundary between the S1 and S2 subunits, which then remain non-covalently bound in the prefusion conformation. S is then further cleaved by host proteases at the S2′ site located immediately upstream of the fusion peptide which is believed to activate the protein for membrane fusion via irreversible conformational changes. Different coronaviruses use distinct domains within the S1 subunit to recognize a variety of attachment and entry receptors, depending on the viral species. SARS-CoV-2 has been demonstrated to bind with the angiotensin-converting enzyme 2 (ACE2) to enter target host cells. SARS-CoV-2 also contains a novel furin cleavage site at the S1/S2 boundary, which is cleaved during biosynthesis. This polybasic cleavage site may help expand SARS-CoV-2 cell and tissue tropism and/or enhance its transmissibility, compared with other CoVs, due to the near-ubiquitous distribution of furin-like proteases and similar reported effects on other viruses, such as influenza (Walls et al., 2020).
REFERENCES
- Ji et al. (2020). Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. J Med Virol. 2020;10.1002/jmv.25682. doi:10.1002/jmv.25682.Li F. (2016). Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol. 3(1):237-261.
- Lu et al. (2015). Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 23(8):468–78.
- Lu R, Zhao X, Li J, et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. S0140-6736(20)30251-8. doi:10.1016/S0140-6736(20)30251-8.
- Su et al. (2016). Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016 Jun; 24(6):490-502.
- Novel coronavirus (2019-nCoV), World health Organisation (WHO), 2020.
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281-292.e6.