Human Coronavirus 229E Nucleoprotein, His-Tag (E. coli)
$640.54 – $2,403.48 excl. VAT
Human coronavirus 229E nucleoprotein is a recombinant HCoV-229E nucleocapsid protein manufactured in E. coli, with C-terminal His-tag.
HUMAN CORONAVIRUS 229E NUCLEOPROTEIN, HIS-TAG (E. COLI)
Human coronavirus 229E nucleoprotein is a recombinant HCoV-229E nucleocapsid protein manufactured in E. coli, with C-terminal His-tag.
PRODUCT DETAILS – HUMAN CORONAVIRUS 229E SPIKE GLYCOPROTEIN (S1)
-
- Human coronavirus 229E nucleoprotein, corresponding to amino acids 1-389 (NP_073556.1; sequence strain Inf-1 ).
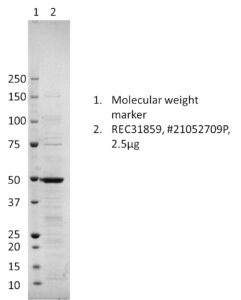
- Protein contains a C-terminal His-tag with apparent molecular weight of 49 kDa.
- Protein was produced in E. coli and purified from culture supernatant by immobilized metal affinity chromtatography, followed by dialysis.
- Presented as liquid, in 20mM sodium phosphate, 25mM potassium carbonate, 150mM NaCl pH10.5.
BACKGROUND
Coronaviruses (CoV) are enveloped, positive stranded RNA viruses in the order Nidovirales, family Coronaviridae, subfamily Coronavirinae. The subfamily Coronavirinae contains the four genera Alpha-, Beta-, Gamma-, and Deltacoronavirus. Alpha- and betacoronaviruses mainly infect humans (Corman et al, 2018). The alphacoronavirus HCoV-NL63 was first isolated in 2003 from a 7-month-old child suffering from bronchiolitis and conjunctivitis (van der Hoek et al., 2004). Infection with the virus has been confirmed worldwide, and has an association with many common symptoms and diseases. Associated diseases include mild to moderate upper respiratory tract infections, severe lower respiratory tract infection, croup and bronchiolitis. The virus is found primarily in young children, the elderly, and immunocompromised patients with acute respiratory illness. It also has a seasonal association in temperate climates. Bat-CoVs have been identified which are related to HCoV-NL63. The genome organization of the NL63-related bat CoVs are similar to that of HCoV-NL63 (ORF1ab-S-ORF3-E-M-N) with the exception that the bat CoV genomes encode an additional open reading frame (ORF X) downstream of the N gene (Tao et al., 2017). However, the zoonotic sources of HCoV-NL63 are unknown. It is thought that HCoV-NL63 may have diverged from a HCoV-229E ancestor in the past, followed by a separation into two distinct HCoV-NL63 lineages. There is also evidence of recombination between viral isolates during the evolution of HCoV-NL63, resulting in a mosaic genome structure (Pyrc et al., 2006). There has been a high occurrence of co-infections reported with HCoV-NL63 and other respiratory viruses, including other human coronaviruses, influenza A virus, respiratory syncytial virus (RSV), parainfluenza virus and human metapneumovirus (hMPV), resulting in increased severity of symptoms and increased opportunities for recombination (Abdul-Rasool & Fielding, 2010).
HCoV-NL63 has a single-stranded RNA genome (27,553 bp) which encodes for a 5′ replicase polyprotein (ORF1a and ORF1b) which encode for the enzymes required for viral RNA replication. The genome also encodes for the 3′ structural proteins, including spike (S), envelope (E), membrane (M) and nucleocapsid (N), which are common to all coronaviruses. The structural proteins are involved in various viral processes, including virus particle formation (Pyrc et al., 2004). Coronavirus spike proteins mediate attachment to the cellular receptors and subsequent fusion of the virus and cell membrane. The N-terminal portion of HCoV-NL63 spike protein contains a unique 179 amino acid domain not present in other coronaviruses. This region represents the most variable region of the HCoV-NL63 genome and a role in immune evasion for this region has been proposed (Pyrc et al., 2007). HCoV-NL63 uses receptor angiotensin converting enzyme (ACE)-2 for entry into the host cell. During virus replication, a variety of proteins are synthesised including the viral RNA binding protein, nucleoprotein (N or nucleocapsid protein), a multi-functional protein with roles in both the virus life cycle and modulating host cell function. One of the key functions of the N protein is to bind viral RNA and selectively incorporate it into virus particles. The genome RNA is complexed with the basic nucleocapsid (N) protein to form a helical capsid found within the viral membrane. The nucleocapsid proteins of coronaviruses representing groups I, II, and III have been shown to localize to the nucleolus as well as to the cytoplasm suggesting that N protein induces a cell cycle delay or arrest, most likely in the G2/M phase, possibly by inhibition of cytokines (Weiss and Navas-Martin, 2005).
REFERENCES
- Abdul-Rasool S, Fielding, BC. Understanding Human Coronavirus HCoV-NL63. Open Virol J. 2010; 4: 76–84.
- Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and Sources of Endemic Human Coronaviruses. Adv Virus Res. 2018;100:163-188.
- Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121(1):190-193.
- Pyrc K, Jebbink MF, Berkhout B, van der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol J. 2004;1:7.
- Pyrc K, Dijkman R, Deng L, et al. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J Mol Biol. 2006;364(5):964-973.
- Pyrc K, Berkhout B, van der Hoek L. The novel human coronaviruses NL63 and HKU1. J Virol. 2007;81(7):3051-3057.
- Tao Y, Shi M, Chommanard C, et al. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J Virol. 2017;91(5):e01953-16.
- van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med. 2004;10(4):368-373.
- Weiss and Navas-Martin (2005). Coronavirus Pathogenesis and the Emerging Pathogen Severe Acute Respiratory Syndrome Coronavirus. Microbiol Mol Biol Rev. 69(4): 635–664.