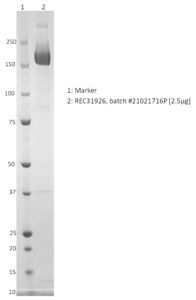
SDS-PAGE: Coomassie-stained reducing SDS-PAGE showing purified full-length spike glycoprotein.
SARS-CoV-2 (B.1.351/501.V2) Stabilized Spike Glycoprotein (Full-Length), His-Strep-Tag (HEK293)
Price range: $1,048.93 through $3,989.68 excl. VAT
SOUTH AFRICAN (BETA) VARIANT
SARS-CoV-2, previously known as the 2019 Novel Coronavirus (2019-nCoV), causes the pandemic COVID-19 disease. The B.1.351/501.V2 variant, also known as 501.V2, 20H/501Y.V2 (formerly 20C/501Y.V2), was first detected in South Africa and includes the amino acid substitutions D80A, D215G, K417N, E484K, N501Y and A701V relative to the prototypic Wuhan-Hu-1 strain.
SARS-COV-2 (B.1.351/501.V2) STABILIZED SPIKE GLYCOPROTEIN (FULL-LENGTH), HIS-STREP-TAG (HEK293)
SARS-CoV-2 (501.V2) stabilized spike glycoprotein, from the South Africa variant. SARS-CoV-2, previously known as the 2019 Novel Coronavirus (2019-nCoV), causes the pandemic COVID-19 disease.
PRODUCT DETAILS – SARS-COV-2 (501.V2) STABILIZED SPIKE GLYCOPROTEIN (FULL-LENGTH), HIS-STREP-TAG (HEK293)
- SARS-CoV-2 (B.1.351/501.V2) stabilized spike, a full-length spike glycoprotein (“South African variant”)
- SARS-CoV-2 HexaPro stabilized spike contains deletions and amino acid changes in accordance with variant B.1.351/501.V2 (“South African variant”): D80A, D215G, K417N, E484K, N501Y, A701V relative to Wuhan-Hu-1
- This protein contains 6 amino acid changes to proline and a C-terminal T4 fibritin trimerization domain to stabilize the protein in its pre-fusion conformation as described in Hsieh, Goldsmith et al. 2020.
- Expressed in HEK293 and purified by affinity chromatography.
- Presented in DPBS at
BACKGROUND
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus that causes coronavirus disease 2019 (COVID-19). The sequence WIV04/2019, belonging to the GISAID S clade / PANGOLIN A lineage / Nextstrain 19B clade, is believed to be the original sequence infecting humans (Zhukova et al., 2020). However, there are many thousands of variants of SARS-CoV-2 (Koyama et al., 2020) and subtypes of the virus can be placed into much larger groupings such as lineages or clades.
The 501.V2 variant, also known as 501.V2, 20H/501Y.V2 (formerly 20C/501Y.V2), or lineage B.1.351, was first detected in South Africa in December 2020 (Tegally et al., 2020). It was reported that the prevalence of the variant was higher among young people with no underlying health conditions, more frequently resulting in serious illness compared to other variants. It was speculated that this variant may be driving a second wave of the COVID-19 epidemic in the country due to it spreading at a more rapid pace than other earlier variants of the virus. Routine sequencing by South African health authorities found that this new SARS-CoV-2 variant had largely replaced other SARS-CoV-2 viruses circulating in the Eastern Cape, Western Cape, and KwaZulu-Natal provinces. The 501.V2 variant contains several mutations that allow it to attach more easily to human cells, namely N501Y, K417N, and E484K and preliminary studies suggest the variant is associated with a higher viral load, which may suggest potential for increased transmissibility. The N501Y mutation is also present in the UK Lineage B.1.1.7. This mutation is of concern because it is located in the SARS-CoV-2 (501.V2) spike RBD, the viruses receptor-binding domain and increases binding to the receptor ACE2 (angiotensin converting enzyme 2).
REFERENCES
- Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020 Jul 1;98(7):495-504.
- Tegally, H. et al. Preprint at medRxiv https://doi.org/10.1101/2020.12.21.20248640 (2020).
- WHO. SARS-CoV-2 Variants. Disease Outbreak News. 31 December 2020
- Zhukova A, Blassel L, Lemoine F, Morel M, Voznica J, Gascuel O. Origin, evolution and global spread of SARS-CoV-2. C R Biol. 2020 Nov 24. doi: 10.5802/crbiol.29.