SARS-CoV-2 Non-Structural Proteins
In addition to antigens, antibodies and cell-surface receptors, The Native Antigen Company offers various recombinant SARS-CoV-2 non-structural proteins (Nsps). These Nsps can be used in a broad variety of research and drug development applications. Some of these proteins are biologically active enzymes, as determined by enzymatic and inhibiting biochemical assays. To facilitate purification and binding in assays, these proteins also include C- or N-terminal his-tags. Click the buttons below for more information, or click here to see our full coronavirus range.
Nsp1: Host Shutoff Factor
The viral nonstructural protein 1 (Nsp1) is the only membrane-associated protein that anchors the replication complex to the cellular membranes. Nsp1 inhibits host translation by interacting with the 40S ribosomal subunit. The Nsp1-40S ribosome complex further induces an endonucleolytic cleavage near the 5’UTR of host mRNAs, targeting them for degradation. Viral mRNAs are not susceptible to Nsp1-mediated endonucleolytic RNA cleavage thanks to the presence of a 5′-end leader sequence and are therefore protected from degradation. By suppressing host gene expression, Nsp1 facilitates efficient viral gene expression in infected cells and evasion from host immune response (Lapointe et al., 2021).

SDS-PAGE: Coomassie-stained reducing SDS-PAGE showing purified Nsp1.
Nsp3: Papain-Like Protease (PLpro)
SARS-CoV-2 papain-like protease (PLpro) is a cysteine protease that acts on three cleavage sites of the viral polyprotein to release mature non-structural proteins (NSP) 1, 2 and 3 (Kandeel et al., 2020). PLpro also strips ubiquitin and ISG15 from host-cell proteins to help coronaviruses to evade the host innate immune responses. PLpro is therefore involved in inhibiting the production of cytokines and chemokines, that are responsible for the activation of the host innate immune response against viral infection. Consequently, PLpro is an important molecular target in the design of various different SARS-CoV-2 antivirals.
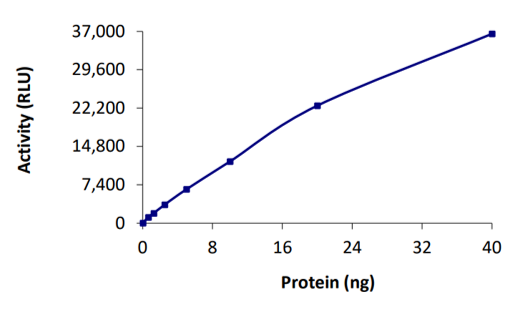
DUB-specific activity of SARS-CoV-2 PLpro at different concentrations.
Nsp5: 3C-Like Protease (3CLpro)
SARS-CoV-2 3C-like proteinase (3CLpro) is the 5th non-structural protein (NSP5) that uses cysteine activity to cleave the C-terminus of the large replicase polyprotein, 1ab at no fewer than 11 sites (Zhang et al., 2020). The functional importance of 3CLpro in the viral life cycle makes this protease an attractive target for the development of drugs directed against SARS-CoV-2.
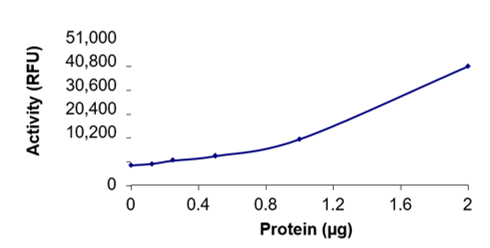
The protease specific activity of 3CLpro was determined to be 4170 pmol/min/mg at an enzyme concentration of 1.2 µM.
Nsp7: RdRp Subunit
Nsp7 and Nsp8 stabilize regions of Nsp12 (RNA-dependent RNA polymerase; RdRp) involved in RNA binding and are essential for a highly active Nsp12 polymerase complex. Nsp7 has been suggested to confer RNA-binding properties to the trimeric polymerase complex (Subissi et al., 2014).
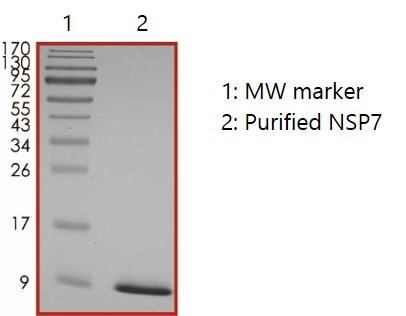
SDS-PAGE: Coomassie-stained reducing SDS-PAGE showing purified Nsp7.
Nsp8: RdRp Subunit
Nsp7 and Nsp8 stabilize regions of Nsp12 (RNA-dependent RNA polymerase; RdRp) involved in RNA binding and are essential for a highly active Nsp12 polymerase complex. Nsp8 has been shown to act as an oligo(U)-templated polyadenylyltransferase and a robust (mono/oligo) adenylate transferase (Tvarogova et al., 2019). Amino acid substitutions in RdRp (P323L) have been shown to cause conformational changes near the Nsp8 binding site that may affect virus replication and transcription (Alosaimi et al., 2021).
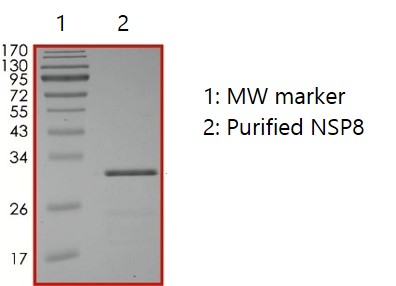
SDS-PAGE: Coomassie-stained reducing SDS-PAGE showing purified Nsp8.
Nsp10/16: Methyltransferase (active)
Many eukaryotic viruses have evolved 2′-O-methyltransferases (2′-O-MTase) to modify their viral mRNAs and carry a cap-1 structure (m7GpppNm) at the 5′ end. This 5’ cap structure is important for viral mRNA stability, protein translation and viral immune escape. SARS-CoV possess Nsp16 that has 2′-O-MTase activity. Nsp16 requires Nsp10 for activation which is a conserved mechanism in corona viruses. Mutations have been identified in SARS-CoV-2 Nsp10 and Nsp16, which alter Nsp10/Nsp16 secondary structure, and protein-modelling studies have revealed that such mutations can affect the proteins dynamicity and stability (Azad et al., 2020). Therefore, inhibitors targeting the Nsp10/Nsp16 2′-O-MTase are potential targets for developing anti-coronavirus drugs (Chen et al., 2011).
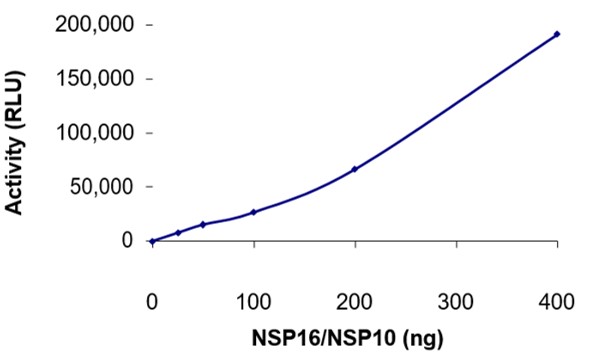
ELISA: The specific activity of SARS-CoV-2 NSP10/NSP16 methyltransferase was determined to be 53 pmol/min/mg (Methyltransferase-Glo™ Methyltransferase Assay, Promega).
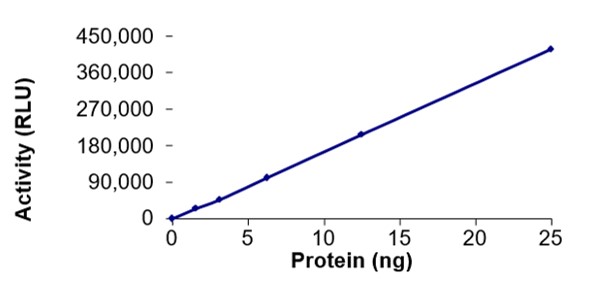
Nsp12: RdRp Catalytic Subunit
RNA-dependent RNA polymerase (RdRp) is an enzyme that is crucial to life cycle of RNA viruses including coronaviruses, catalysing the synthesis of viral RNA in complex with Nsp7 and Nsp8 (Gao et al., 2020). Given its central role in the coronavirus life cycle, RdRp is a prime target for nucleotide analog inhibitors, such as remdesivir.
RdRp activity determined by detection of pyrophosphate (PPi) generated during nascent strand synthesis by a chemiluminescent coupled assay method using ATP sulfurylase and luciferase.
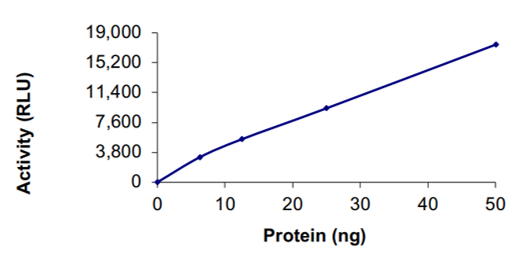
Nsp14: Methyltransferase (active)
Coronaviruses codes for a bifunctional non-structural protein 14 (NSP14) that is important for viral replication and transcription. Nsp14 contains an N-terminal exo-ribonuclease (ExoN) domain for proof reading during viral replication and a C-terminal N-7 methyltransferase (N7-MTase) domain for mRNA capping. Nsp14-ExoN is required for native recombination, and inactivation of ExoN results in decreased recombination frequency and altered recombination products (Gribble et al., 2021). Nsp14 associates with several viral proteins. It can bind to Nsp10 and the resulting complex exhibits enhanced ExoN activity in vitro. It can also bind to the polymerase complex (Nsp12/Nsp8/Nsp7) forming a complex that retains all associated enzymatic activities. Nsp14 is known to be a good target for potential small molecule inhibitors against SARS-CoV-2 (Umar et al., 2021).
ELISA: The specific activity of SARS-CoV-2 NSP14 methyltransferase was determined to be 14.1 nmol/min/mg (Methyltransferase-Glo™ Methyltransferase Assay, Promega).
Our Coronavirus Range
The Native Antigen Company offers an extensive and growing range of SARS-CoV-2 antigens, suitable for a variety of applications, including assay development, vaccine screening, neutralisation testing, immunofluorescence, surface plasmon resonance and academic research. We also offer a variety of monoclonal and polyclonal coronavirus antibodies for use in immunoassays, Western blots, neutralisation assays, surface plasmon resonance and X-ray crystallography. Anti-SARS-CoV-2 Spike (CR3022) IgG antibodies are available in both humanised and rabbit-derived forms, in addition to a humanised IgM. An anti-Spike IgA monoclonal with affinity for the SARS-CoV-2/SARS-CoV RBD is also available.
The Native Antigen Company also offers an extensive and growing range antigens for SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-NL63, HCoV-229E and HCoV-HKU1. These antigens are suitable for a variety of applications, including assay development, vaccine screening, neutralisation testing, immunofluorescence, surface plasmon resonance and academic research.
