RT-PCR is the workhorse of viral diagnosis and has been invaluable in COVID-19 case confirmation and isolation guidance. However, while fast and sensitive, PCR suffers from some inherent drawbacks that limit it to diagnosis during the acute phase of infection. To better understand the transmission dynamics of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and develop effective countermeasures against it, antigen- and antibody-based immunoassays will be essential. In this blog, we explain the key differences between PCR and immunoassays for COVID-19 diagnosis, and present our growing pipeline of coronavirus reagents for the development of highly accurate diagnostic kits.
Viral Diagnostics: A Quick Recap
The two major methods for diagnosing viral infection are the polymerase chain reaction (PCR), and immunoassays:
PCR
The polymerase chain reaction is a routine laboratory technique used to amplify small samples of DNA into larger quantities that can be detected and analysed. For patient diagnosis, a viral RNA or DNA sample is taken by swab or blood draw, before being sent to a specialist laboratory for analysis.
For RNA viruses, such as SARS-CoV-2, the polymerase chain reaction is preceded by an additional step to produce a complementary DNA template (cDNA) from RNA, by the addition of a reverse transcriptase enzyme (hence, Reverse Transcription-PCR (RT-PCR)).
PCR then begins by the addition of short DNA sequences known as primers, which bind the viral DNA strands. The double-stranded section of DNA is then recognised and bound by a thermostable polymerase enzyme which acts as a molecular photocopier to extend the sequence and produce a full, complementary strand.
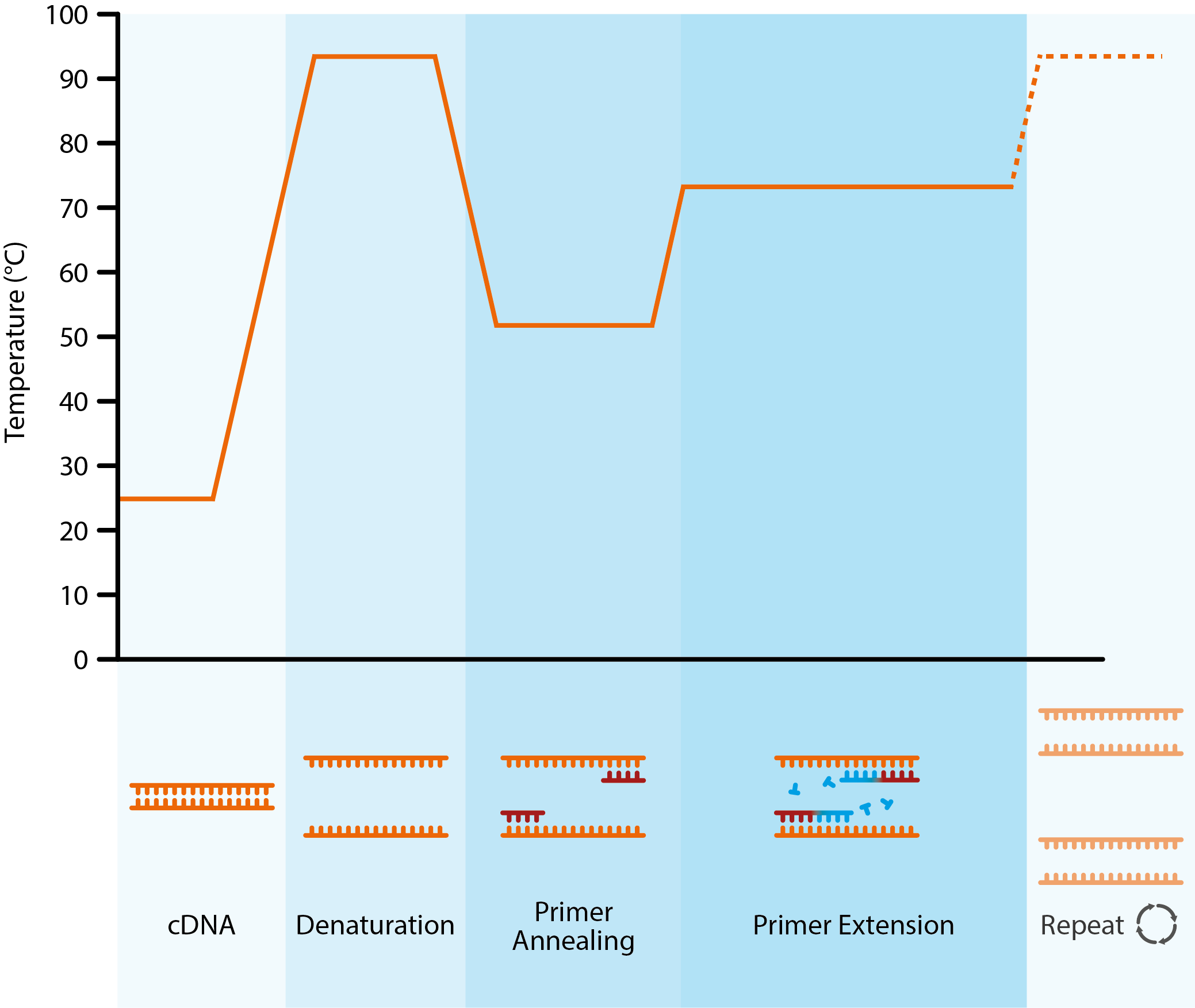
Diagram showing the key stages of the polymerase chain reaction. Once generated, a cDNA template is added to a thermal cycler, along with free nucleotides, primers and Taq polymerase. The dsDNA is denatured with heat to create single strands. The temperature is then lowered to allow primers to anneal to complementary DNA sequences. By raising the temperature once more, the thermostable Taq polymerase can catalyse the addition of nucleotides to growing, complementary DNA strands.
By controlling the annealing, extension and denaturation steps with changes in temperature, the initial sample of viral DNA can be exponentially amplified, followed by the addition of specific DNA probes that produce a detectable signal (often fluorescent) to confirm the etiological agent.
Immunoassays
Unlike molecular techniques, immunoassays detect the presence of specific immune proteins. These assays take on a wide range of different formats, but essentially consist of an antigen or antibody, immobilised on a surface (most often a titre plate or paper strip), which binds virus-specific antigens or antibodies from a patient sample (i.e. sputum or blood sera). By adding a further reporter protein, it is then possible to detect a virus-specific immune signal to confirm the presence of ongoing or past viral infection.
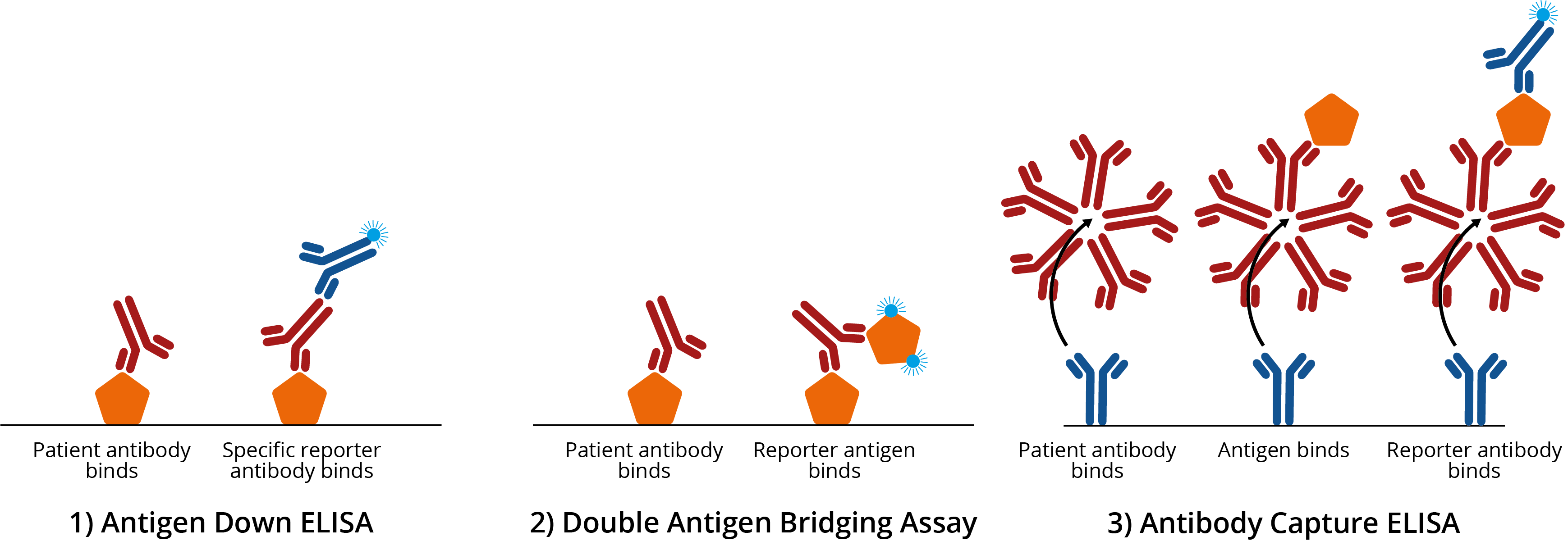
Common examples of enzyme-linked immunosorbent assays (ELISAs). 1A) Antigen down ELISA. Titre plate is coated with antigen, which binds antigen-specific IgG antibodies from patient sera. Addition of a specific reporter antibody then confirms the presence of antigen-specific antibody; 1B) Double antigen bridging assay (DABA). Titre plate is coated with antigen, which binds antigen-specific IgG antibodies from patient sera. A second reporter antigen is then added to create an ‘antigen sandwich’ ; 1C) Antibody capture ELISA. Titre plate is coated with specific antibody, which binds IgM from patient sera. Antigen is then added, before the addition of a reporter antibody to improve signal:noise. For more information on ELISA design, see our blog on ELISA formats.
PCR for COVID-19
As a disease, COVID-19 shows considerable symptomatic variation. Individuals may present with no symptoms at all, while some may have a mild cough and fever, and an unlucky few may experience severe pneumonia and respiratory failure [1]. To complicate matters, COVID-19’s most common symptoms strongly overlap with those presented by other co-circulating respiratory illnesses:
Table comparing the typical symptoms of COVID-19, with the common cold, flu and allergy.
Because of this, diagnosing COVID-19 on the basis of clinical symptoms alone is highly inaccurate and must be confirmed by the use of highly specific diagnostic tests. Fortunately, thanks to rapid sequencing and publication of the SARS-CoV-2 genome in early January [2], RT-PCR primers and open access protocols were made quickly available, and are now being used by medical facilities worldwide to diagnose patients.
The main advantage of RT-PCR has been its speed and sensitivity. After taking a pharyngeal swab from the back of a patient’s throat, a sample can be sent to the lab to provide results within hours [3]. And as only a very small amount of viral RNA needs to be present for amplification, these tests are highly sensitive in detecting virus from a sample.
However, RT-PCR diagnosis of COVID-19 has its limitations. Detecting SARS-CoV-2 from pharyngeal swabs requires high-quality specimens that contain a sufficient amount of intact viral RNA. Yet, SARS-CoV-2 loads in the respiratory tract have shown to vary considerably. This has not only led to high false-negative rates, with probable cases remaining negative after multiple swabs, but is further exposing healthcare workers to risk of infection [3][4].
There is also an issue of scale. Processing of COVID-19 samples requires specialised biocontainment laboratories, operated by highly trained technicians, usually only found within medium-large hospital facilities [5]. While capacity is normally sufficient, the COVID-19 pandemic is pushing these facilities to their limit.
For more resource-limited countries, diagnostic infrastructure and training is even harder to come by, with many healthcare systems having only a handful of labs and technicians spread across vast geographical areas. Governments and private organisations are working fast to increase the capacity and speed of PCR testing, but as laboratories are strained by growing case numbers, delays and complete lack of testing is becoming all-too-common [6].
Where do Immunoassays Fit In?
At a glance, immunoassays show some distinct advantages over PCR. Antigens and antibodies are considerably more stable than RNA [7], which makes them less susceptible to spoliation during transport and storage, therefore reducing the chance of false-negative results. Testing accuracy is also improved by the fact that antigens and antibodies are more uniformly available in sputum and blood samples.
However, the biggest advantage of immunoassays is their ability to detect past infections.
Once a patient has recovered from COVID-19 and virus is cleared from the body, viral RNA is no longer available for detection in the respiratory tract, leaving only a short window during the acute stage of infection in which SARS-CoV-2 can be detected. While this works well for the diagnosis of ongoing infections, it gives no indication of whether a patient has had the infection historically, and what their immune status is (i.e. if they are immune to COVID-19 or still susceptible to infection).
Unlike RNA, antibodies are long-lasting and can persist in the bloodstream for many years after infection. As such, immunoassays enable us to identify patients that have had COVID-19, retrospectively. The type of antibody and its relative levels could also be used to indicate the stage of infection and estimate time since exposure for contact tracing.
However, antibody tests have their limitations too. As immunological data continues to emerge, it is becoming apparent that the body’s antibody response to COVID-19 is slow – considerably slower than we might expect. While data at this point is still limited, it appears that the initial IgM antibody response doesn’t peak until ~9 days after initial infection and the IgG antibody response doesn’t peak until day ~11 [8][9].
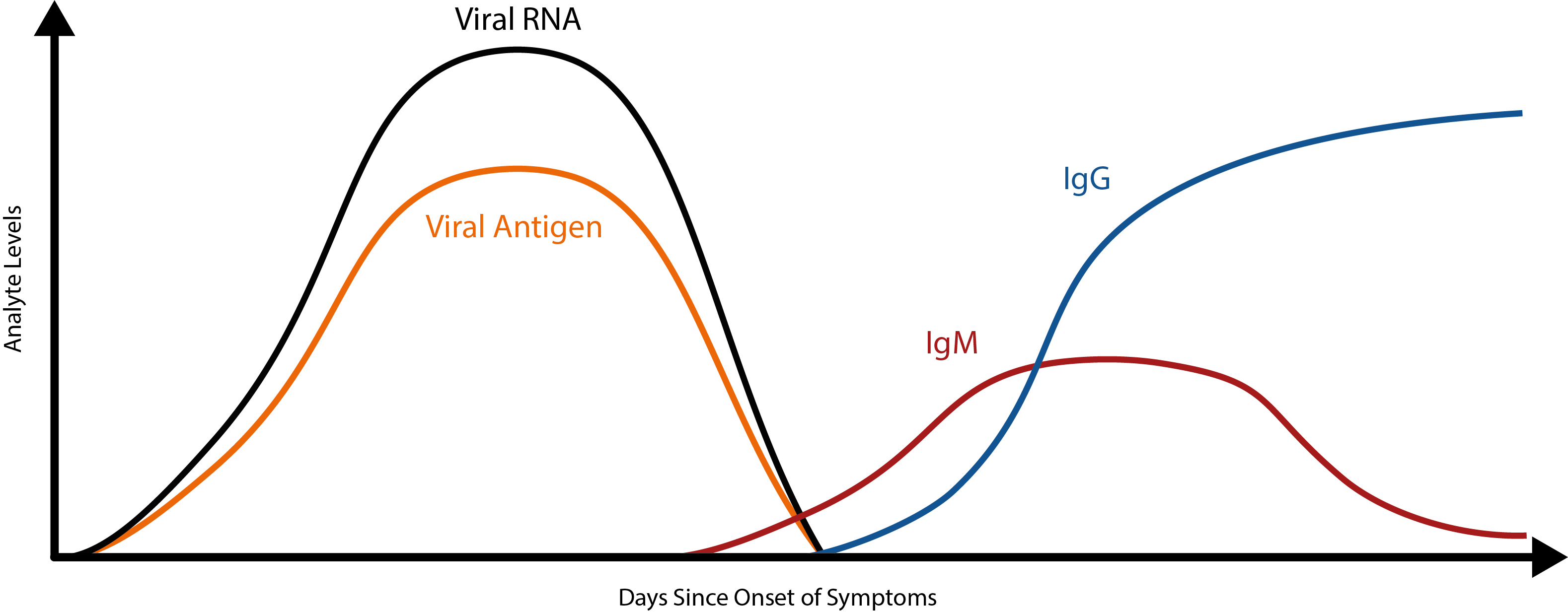
To put this into perspective, most viruses elicit a primary IgM response within 5 days [10]. Because of this, SARS-CoV-2 antibodies are unlikely to make good markers of acute COVID-19 infection. While early studies have shown that combined RT-PCR/antibody testing can reduce false-negative rates [11], the use of antibody kits for acute-phase diagnosis alone makes for a risky strategy.
So where does this leave antibody testing?
While antibodies may not be appropriate for acute-phase diagnosis, they still show many valuable applications for COVID-19. Potentially the most valuable use of wide-scale antibody testing is as a public health tool.
To make the right public health decisions, governments worldwide are using estimates of transmission rates, case numbers and case fatality rates (CFR). However, given that anywhere from 20-80% of COVID-19 cases are estimated to be asymptomatic [12], these figures have been very difficult to accurately model. On a population level, this means that the true size and scope of the pandemic is still undefined, leaving policy makers with little indication of how serious of a threat COVID-19 still is, and how long it can be expected to last.
By conducting random antibody sampling of the general public (known as a serosurvey), public health bodies could better estimate the true levels of exposure and resulting population immunity. For COVID-19, this would be a game-changer, as true transmission and CFRs could be calculated to forecast the intensity and longevity of the pandemic to direct decision-making. Furthermore, by identifying potential geographical ‘hot-spots’ of low population immunity, health systems could better allocate resources to prevent or manage transmission.
Testing at the Point of Care
While antibodies aren’t suitable for diagnosing active COVID-19 infections, antigens are likely detectable in the sputum from the onset of symptoms [13]. What’s more, by combining antigen detection with a format that can provide rapid results for use at the point-of-care (PoC), community-based, or even door-to-door testing could soon become a reality – allowing those who suspect an infection to get quick results and determine whether they should isolate and/or seek medical care.
The major caveat of formats such as ELISA, are that they are unwieldy, and require trained scientists, operating in sterile laboratory conditions – not to mention, the complications of specimen collection and cold-chain logistics, that use up scarce healthcare resources.
Fortunately, a range of PoC formats have been developed with simplicity and portability in mind. So-called lateral flow assays (LFAs) are the most popular choice. These simple and easy-to-produce devices rely on the property of fluid analytes to move through absorbent substances on their own accord, via a phenomenon known as capillary flow.
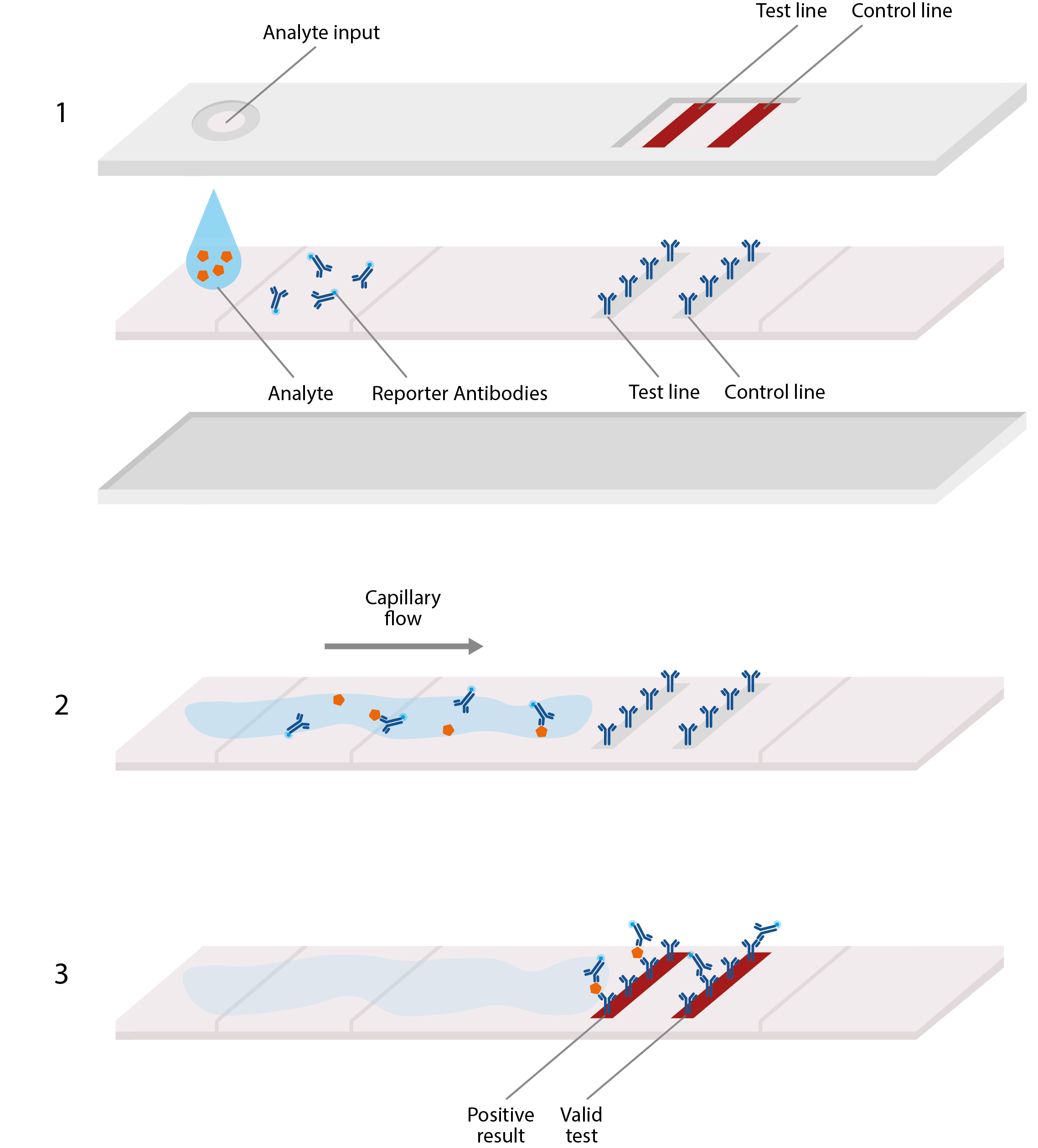
Basic design of lateral flow assay, akin to home pregnancy tests. LFAs typically comprise a plastic cassette, containing a strip of paper that is able to absorb and transport analyte. 1) Analyte from blood or sputum is first added to one end of the paper strip, where it is absorbed by the paper, allowing the viral antigens to migrate along it. 2) These antigens first encounter an area of conjugate antibodies, which adhere to them, along with detectable tags. 3) The antigens then migrate to an area of test antibodies that are highly specific to the virus and bind to produce a solid, visible line that indicates a positive result, and hence, confirmed infection. Secondary control lines can then be used to detect conjugate antibodies and ensure that the test is working correctly.
These tests would be invaluable in providing rapid diagnosis in local communities, to give both healthcare systems and public health bodies a much clearer picture of unfolding pandemic.
Once governments are able to start reducing transmission, and COVID-19 case numbers begin to fall, PoC tests will become even more crucial in identifying asymptomatic carriers and infected individuals to ensure they are isolated from the general population.
While these kits are not currently available for widespread use, both public and private organisations, worldwide are working on prototypes, with over 50 currently in development [14].
Developing a Vaccine
So far, there are over 40 vaccine candidates for COVID-19 currently in the pipeline, with Moderna Therapeutic’s front-runner set to start human trials as early as next month [15]. While these vaccines are based on a wide range of platforms, (including mRNA, DNA, nanoparticles, subunits, synthetic peptides and virus-like particles, to name a few [16]), it can be said with near certainty, that a SARS-CoV-2 vaccine will elicit immune responses to the Spike protein.
Spike proteins are large, type I transmembrane proteins that protrude from the surface of the coronavirus virion. These proteins are immunodominant (i.e. induce the strongest immune response), with the ability to elicit high neutralising antibody titres (i.e. antibodies that bind to the virus and prevent it from entering cells) [17].
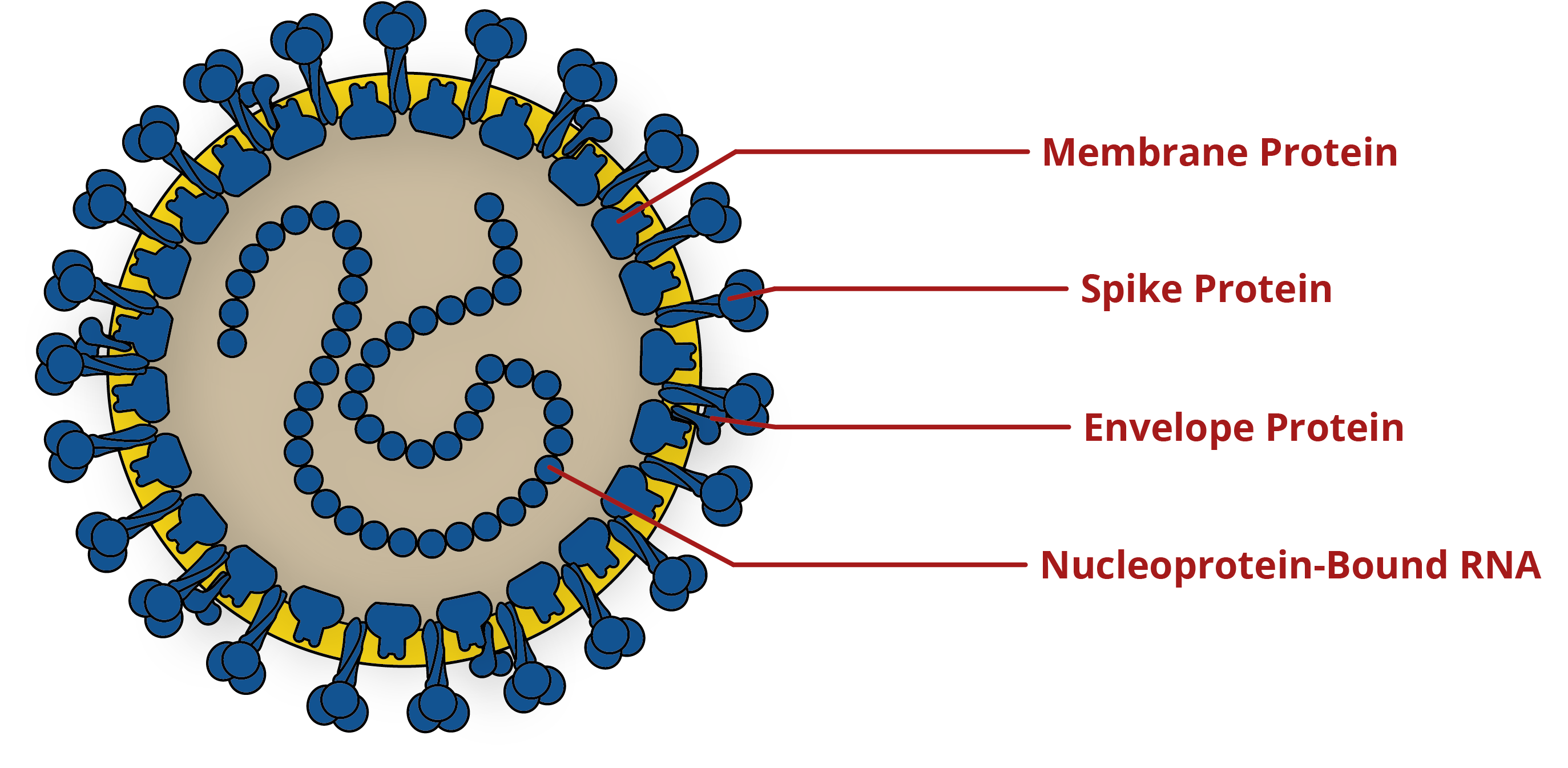
Diagram of coronavirus, showing key antigens for the development of diagnostics and vaccines. For more information on the Spike protein, please see our blog on its structure and immunology.
As such, administration of a successful SARS-CoV-2 vaccine will induce anti-Spike IgG and IgM antibody responses to confer protection. However, serologically-speaking, vaccine-induced antibodies may be indistinguishable from natural infection.
This creates a headache for vaccine manufacturers, who need to be able to differentiate and measure vaccine-specific immune responses to prove efficacy during clinical trials. Therefore, alternative antigens will need to be used when designing assays, to be able to differentiate between vaccine-induced and natural immune responses.
While there are multiple antigens to choose from, Nucleoprotein is a strong contender. The Nucleoprotein is a vital structural protein with the primary function of forming a complex with viral RNA to mediate packaging and replication. While not as immunodominant as Spike, it is highly immunogenic and is profusely over-expressed during infection [18].
The challenge in using Nucleoprotein, however, is its similarity between coronavirus strains. Unlike Spike, Nucleoprotein shows less genetic variation – especially between SARS-CoV-2 and the genetically distinct, 2002 SARS Coronavirus. The problem here, is that antibodies to SARS-CoV Nucleoprotein have the potential to cross-react with SARS-CoV-2 Nucleoprotein (i.e. the same antibodies are able to bind Nucleoprotein from both coronaviruses). Indeed, studies have shown the coronavirus Nucleoprotein to be broadly cross-reactive [19][20].
Consequently, immunoassay developers will need to thoroughly screen the SARS-CoV-2 Nucleoprotein for antibody binding, and consider designing innovative immunoassay formats to avoid generating false-positive results.
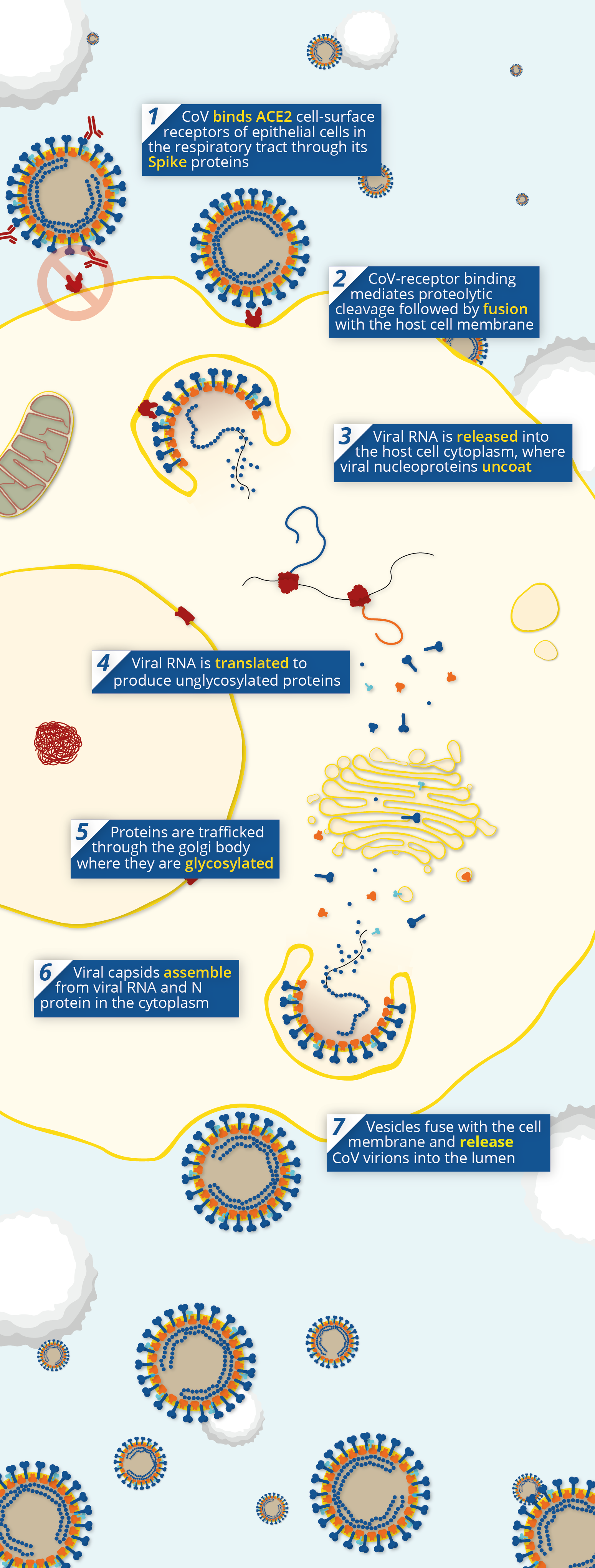
Pathway of coronavirus pathogenesis, showing the point at which neutralising antibodies prevent viral entry via Angiotensin-Converting Enzyme 2 (ACE2).
The Convalescent Sera Option
With a vaccine still some time away, intermediary measures are being explored to protect those most at risk of severe COVID-19 infection. One of these is passive antibody therapy – a technique in which blood is taken from a recovered patient and purified for administration to protect or treat individuals.

Extraction, purification and administration of convalescent patient sera as a COVID-19 therapy. Adapted from Casadevall and Pirofski.
The antibodies conferred from the donor’s blood act as a stop-gap to provide immediate immunity while the individual develops their own humoral response to fight the infection. The main mechanism for this is through the action of neutralising antibodies, but also includes antibody-dependent cellular cytotoxicity (ADCC) and antibody-mediated phagocytosis.
Passive antibody therapy dates back to the 1918 influenza pandemic, and was used with great effect during the recent 2009 H1N1 Influenza and 2013/2014 Ebola epidemics, showing significantly longer survival in treated patients [21]. For COVID-19, antibodies could be used both prophylactically, to protect healthcare workers in COVID-19 wards, as well as a therapeutic for patients with severe infection.
To develop viable sera, patients’ blood will be need to be screened for anti-SARS-CoV-2 antibodies (preferably neutralising). This is most commonly done with so-called plaque neutralisation assays (PRNT). However, PRNT throughput is limited and needs to be carried out in specialised biocontainment facilities. Therefore, immunoassays will likely have a crucial role to play in the qualitative and quantitative screening of convalescent sera.
Coronavirus Antigens and Antibodies from The Native Antigen Company
The Native Antigen Company offers an extensive and growing range of reagents for the human coronaviruses, including the emergent SARS-2, SARS, MERS coronaviruses, as well as the OC43, HKU1, 229E, and NL63 endemic coronaviruses.
In February 2020, The Native Antigen Company became one of the world’s first suppliers to release commercially-available SARS-CoV-2 antigens. Since then, our scientists have been hard at work, extending our coronavirus range to support the research and development of urgently-needed diagnostics and vaccines.
Our antigens are produced using our proprietary, VirtuE (HEK293) expression system, which ensures full glycosylation and proper folding, to ensure full biological and antigenic activity. To see our product range, please click on the available products below. You can keep up-to-date with new product developments on our Coronavirus Pipeline page.
If we are not currently developing something that you’re interested in, please get in touch at contact@thenativeantigencompany.com, as we may be able to develop it for you.
References
1. https://link.springer.com/article/10.1007/s11606-020-05762-w
2. https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/
3. https://www.nature.com/articles/d41587-020-00010-2
4. https://www.preprints.org/manuscript/202003.0184/v1
5. https://hbr.org/2020/02/we-need-a-cheap-way-to-diagnose-coronavirus
6. https://www.vox.com/2020/3/20/21188266/coronavirus-test-us-united-states
7. http://book.bionumbers.org/how-fast-do-rnas-and-proteins-degrade/
8. https://www.medrxiv.org/content/10.1101/2020.03.06.20031856v1
9. https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.25727
10. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3820351/
11. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3546052
12. https://science.sciencemag.org/content/early/2020/03/13/science.abb3221
13. https://jcm.asm.org/content/42/7/2884.short
14. https://www.finddx.org/covid-19/pipeline/
16. https://www.who.int/blueprint/priority-diseases/key-action/novel-coronavirus-landscape-ncov.pdf?ua=1
17. https://www.biorxiv.org/content/10.1101/2020.01.28.923011v1
18. https://pubmed.ncbi.nlm.nih.gov/12584316/
19. https://www.nature.com/articles/s41586-020-2012-7
20. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC404591/
21. https://www.jci.org/articles/view/138003